Abstract
Halyomorpha halys has recently established in countries outside of its Asian area of origin. In the North Eastern USA, this pest causes severe economic loss in agricultural crops. Breeding populations have been confirmed in Switzerland and Canada; however, their pathways of entry and genetic diversity remain unknown. To determine invasion pathways and source population(s), the diversity of specimens from Asia, North America, and Europe was investigated. Genetic diversity in Asia was higher than that in the Canada and Switzerland. Although three haplotypes were detected among Canadian H. halys, one haplotype dominated. In contrast, two out of three haplotypes were found frequently among Swiss samples. Based on molecular data and interception records it appears likely that H. halys in Canada is derived from the movement of established US populations. Further, North American populations likely originated from the Hebei/Beijing regions of China. The area of origin of the Swiss samples remains unclear. Although the dominant haplotype in Switzerland was consistent with Asian samples collected in the Hebei and Beijing provinces, it was not the dominant haplotype in these regions and further sampling is necessary to its confirm the distribution and abundance in the area of origin. The remaining two haplotypes were unique to Switzerland and no matches were found among our Asian samples. Interestingly, no haplotypes were shared between the North American and Swiss samples, indicating that the two invasions were separate and distinct events and did not result from the movement of goods and materials between USA and Europe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The brown marmorated stink bug (BMSB), Halyomorpha halys (Hemiptera: Pentatomidae), is a pest native to subtropical and temperate areas in East Asia, including China, Japan, Korea, and Taiwan (Hasegawa and Umeya 1974; Hoebeke and Carter 2003). It is reported as a polyphagous pest that feeds on a variety of ornamental plants; fruit trees; field and vegetable crops (Funayama 2003; Leskey et al. 2012a). The first reports of invasive BMSB populations outside of Asia occurred in the USA, where the pest is thought to have established in Pennsylvania in the mid-1990’s (Hoebeke and Carter 2003). Since the initial establishment in Pennsylvania, BMSB has spread to 39 states, with the severity of infestation ranging from occasional detection, to nuisance pest status, to severe pest status (Leskey et al. 2012a, b). The mid-Atlantic States are among the most hardest hit of the invaded regions thus far, with severe economic impacts in fruit trees (e.g., apples and peaches) and field crops (e.g., soybeans and sweet corn) (Nielsen and Hamilton 2009a, b; Nielsen et al. 2011).
In Canada, interceptions of H. halys at various ports of entry across the country began in 1993 from countries including China, Japan, Korea, and the USA, with reports of homeowner finds beginning in the Province of Ontario as of 2010 (Fogain and Graff 2011) and established breeding populations in the field confirmed as of July 2012 (Fraser and Gariepy, unpublished). In Europe BMSB was first officially reported from the canton of Zurich in Switzerland in 2007 (Wermelinger et al. 2008). However, later investigations showed that it was already present in Zurich-Seefeld in May 2004 (personal communication R. Burtscher). In the same year, a single individual was found near Balzers in Lichtenstein, which probably originated from nearby founder populations in Zurich (Arnold 2009). Mueller et al. (2011) suggest that reports of H. halys in private residences in Zurich have increased exponentially between 2007 and 2010, with some economic damage reported in peppers. Outside the Canton of Zurich, H. halys is confirmed to be present in the Cantons Basel-Stadt, Basel-Land, Aargau, Bern, St. Gallen, and Schaffhausen (Wyniger and Kment 2010; Haye and Wyniger 2013). In addition, a single individual was found near Konstanz in southern Germany (Heckmann 2012) and most recently in the Alsace region of France (Callot and Brua 2013). To date, no breeding populations have been found outside Switzerland, but with growing populations a range expansion of H. halys into bordering countries can be expected in the near future.
The exact pathway of entry of BMSB into newly invaded areas is largely unknown. H. halys is considered an excellent hitchhiker, which is often transported from Asia to other locations on inanimate objects as a stowaway in cargo, packing crates, aircraft, machinery, vehicles, and personal luggage (Hoebeke and Carter 2003). Consequently, BMSB has been frequently intercepted by border officials in a number of areas, including Canada (Fogain and Graff 2011), New Zealand (Duthie 2012), Australia (Walker 2009), and Germany (Freers 2012). In New Zealand, H. halys has been intercepted 9 times in both border and post-border inspections between 1999 and 2010, primarily from shipments originating in Japan, but also from containers arriving via the USA, Australia, and Hong Kong (Duthie 2012). Australia does not have an established population of BMSB; however, interceptions from shipments originating from Korea, China, and the USA have been reported (Walker 2009; Australian Department of Agriculture, Fisheries and Forestry 2012a, b). It is suggested that a genetically homogenous population or single mated female was introduced from shipping containers originating from China or Korea, and established and initiated the population outbreak and spread in the USA (Aldrich et al. 2009; Nielsen 2011; Duthie 2012). In Switzerland, it is suspected that H. halys was imported together with woody or floral ornamental plants or fruits (Wermelinger et al. 2008) or in shipping crates and material imported from areas with high populations levels of BMSB (Hoebeke and Carter 2003). The invading population of BMSB in Canada may be derived from a range expansion of the established US population; however, numerous interceptions from Asia have been recorded in Canada and similar arrivals may have resulted in established populations.
Pathway analysis of exotic pests often involves the gathering of shipping/interception information, field trapping data, and genetic data in an effort to assess risks, develop effective exclusion strategies, and improve quarantine policies (Hulme et al. 2008; Barr 2009; Estoup and Guillemaud 2010; Blanchet 2012). Invasion events generally occur over large temporal and spatial scales, and can involve multiple introductions of a small number of individuals from one or more source populations (Corin et al. 2007). Many of these events go undetected, unreported, and/or the data are not readily available to those outside of phytosanitary control organizations (Corin et al. 2007; Piel et al. 2008; Hulme 2009). As such, interception data do not always provide an adequate foundation to reconstruct the invasion history of a given pest species (Estoup and Guillemaud 2010). Molecular resources are well-placed to supplement interception and trapping data to provide a more complete picture regarding the invasion history of a given organism (Miller et al. 2005; Corin et al. 2007; Barr 2009; Estoup and Guillemaud 2010; Blanchet 2012). Accessibility and affordability of molecular technology, coupled with the expansion of databases housing publically available DNA sequence data (e.g., GenBank and the Barcode of Life Database) have led to an increase in the use of molecular tools to evaluate the diversity and potential origin of source populations of invasive species.
The introduced species generally display a loss of genetic diversity during colonization of a new area (Mayr 1954). Analysis of intraspecific variation in mitochondrial DNA provides an effective tool for estimating the phylogeographic history of an organism when individuals are available from both the area of introduction and the area of origin (Avise 1995; Avise et al. 1987). DNA sequence data from mitochondrial genes (e.g., cytochrome b, cytochrome oxidase I, and cytochrome oxidase II) have been successfully used to evaluate the geographic origin and colonization history of a number of introduced invertebrates (Grapputo et al. 2005; Corin et al. 2007; Barr 2009; deLeón et al. 2011; Franks et al. 2011; Teske et al. 2011; He et al. 2012; Auger-Rozenberg et al. 2012; Kanuch et al. 2012; Smith et al. 2012a, b). In particular, the COI gene has demonstrated the ability to separate genetic lineages due to a high level of variability (Stephens et al. 2011; Teske et al. 2011; Smith et al. 2012a, b). To gain a better understanding of the invasion pathways and potential source(s) of H. halys populations in Canada and Switzerland, the mitochondrial cytochrome oxidase I gene (COI) was used to evaluate the genetic diversity and relatedness of introduced and native populations. This information may be useful to determine whether the recently invaded areas were founded by single vs. multiple source populations, and whether genetic diversity levels suggest a founder effect.
Materials and methods
Field collection of the established populations of H. halys
H. halys is highly mobile and is likely to disperse over long distances to overwinter in permanent structures (Watanabe et al. 1994; Cuituan et al. 1993). Often, the first indication of a potential establishment of this species in a newly invaded area is the documentation of homeowner finds when overwintering populations seek shelter within structures in urban areas. Collections of specimens from homes and residential buildings could potentially provide a greater diversity of specimens that have flown in from different, isolated pockets where this species has recently become established in small, localized populations. In contrast, individuals collected on the same or neighboring host plants in a field are more likely to be derived from one adult female, or a small number of related, adult females. As such, representative samples from both residential and field-collected material may capture a greater range of the genetic diversity of H. halys in recently invaded areas in Canada and Switzerland.
Canada: In 2011 and 2012, individual specimens of adult H. halys were obtained from residential locations in Hamilton, Burlington, and Newboro (Province of Ontario), following reports from homeowners indicating their presence. One individual collected from a hotel in Edmonton, Alberta and another individual found in a private residence near Montreal, Quebec were also included. In addition, H. halys nymphs and adults were collected on invasive buckthorn (Rhamnus carthartica (Rhamnaceae)) in natural/wooded habitats in Hamilton, Ontario. Individuals were either directly collected from the foliage by hand, or using a battery-operated aspirator. Alternately, branches were shaken vigorously over a large white sheet, and any H. halys which dropped were subsequently collected using an aspirator.
Switzerland: In the late summer and fall of 2012, individual specimens of H. halys were collected from residential areas in the Cantons of Bern, Zurich, Basel, and Aargau following homeowner reports of their occurrence in the region. Adult H. halys were generally captured by hand or by sweep net on outdoor furniture, siding, balconies, cracks, and crevices of neighborhood buildings. In addition, H. halys nymphs and adults were collected by hand and using sweep nets from a variety of trees and shrubs [e.g., Carpinus betulus (Betulaceae), Campsis grandiflora (Bignoniaceae), Buddleja davidii (Scrophulariaceae), and Sorbus aucuparia (Rosaceae)] in the parkland area surrounding the Chinese garden in Zurich-Seefeld and in private gardens in Zurich-Wollishofen.
China: Specimens from the Hebei province were obtained from two light traps permanently set up at the Langfang Experimental Station for monitoring of insect pests. Samples of H. halys adults were taken from these traps on July 12 and 18, 2012. In the neighboring Beijing province, individual adults were collected by hand from various host plants [e.g., Pyrus betulaefolia (Rosaceae) and Robinia pseudoacacia (Fabaceae)] in orchards at Changping and Haidian in July 2012.
Live H. halys collected in light traps, field habitats, and residential areas were killed and preserved in 95 % ethanol for subsequent molecular analysis. All dead or dried specimens retrieved from private residences were stored in ethanol at −20 °C. The preserved specimens from rearing colonies in the USA (Maryland and Pennsylvania) were also obtained. Table 1 indicates the number of specimens collected from each geographic region.
Intercepted specimens of H. halys
In addition to the collection of specimens from the established populations in North America and Switzerland, specimens sporadically intercepted in Canada (prior to official establishment) and Europe were also obtained to serve as a reference for comparison, and to provide specimens originating from diverse locations where widespread collection efforts were not possible (Table 2). Intercepted specimens may help determine the source of introductions and/or provide a better understanding of potential pathways for invasive species entry.
DNA sequencing
Genomic DNA was extracted from a single leg of each H. halys specimens using a Chelex extraction method (Walsh et al. 1991). This approach does not involve multiple tube transfers (thereby reducing potential for contamination) and contains no filtration steps (thereby permitting recovery of small quantities of DNA from dried or degraded specimens). Briefly, a single leg from each individual insect was placed in a 200-μL well of a 96-well microplate in 100 μL of 5 % solution of Chelex 100 Molecular Grade Resin (Bio-Rad Laboratories) and 2 μL of 20 mg/mL Proteinase K. A negative extraction control containing the Chelex and Proteinase K solutions, but no insect tissue, was included in each microplate. Each microplate was sealed and incubated overnight at 55 °C, followed by 10 min at 99 °C. Samples were centrifuged at 13,000 rpm for 5 min to pellet the Chelex solution, and 50 μL of the supernatant (containing DNA) was transferred to wells in a new plate, taking care not to transfer the Chelex residue along with the sample. Microplates containing the extracted DNA were stored at −20 °C until further analysis. PCRs were performed in a 25 μL volume containing 0.125 μL of Taq Platinum, 2.5 μL of 10× PCR buffer, 1.25 μL of 50 mM MgCl2, 0.125 μL of 10 μM dNTPs (Invitrogen), 0.25 μL of 10 μM forward and reverse primer (respectively), 19.5 μL ddH20, and 1 μL of template DNA.
Cytochrome Oxidase I: A 658-bp sequence of the mitochondrial Cytochrome C oxidase subunit 1 (COI) was amplified by PCR using primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (see Folmer et al. 1994).
28S: A 517-bp fragment of the nuclear 28S gene was amplified as per Kavar et al. (2006) using the forward primer 5′-CCCGTCTTGAAACACGGACCAA-3′ and the reverse primer 5′-CCACAGCGCCAGTTCTGCTTAC-3′.
Cytochrome B: A 426-bp fragment of the mitochondrial Cytochrome B gene was amplified using the primers described by Muraji et al. (2000) (forward: 5′-TAGGATATGTTTTACCTTGAGGACA-3′; reverse: 5′-CTCCTCCTAATTTATTAGGAATTG-3′).
PCR products were subject to electrophoresis on a 1.5 % agarose gel (UltraPure Agarose, Invitrogen) containing 1x GelRed (Biotium). The resulting PCR products were visualized on a BioDoc-it imaging system (UVP) and purified using ExoSAP-IT (USB Corporation) following the manufacturer’s instructions. The PCR products were bi-directionally sequenced (using the above primers) on an ABI 3730 DNA Analyzer at the Robarts Research Institute (London Regional Genomics Centre, ON, Canada). Forward and reverse sequences were assembled, aligned using Clustal W algorithm (Thompson et al. 1994), and edited by using Codon Code Aligner program, version 4.0.4. Amplification and sequencing of specimens which produced unique, singly occurring haplotypes was repeated to ensure that sequence variation was not an artifact generated by errors in the PCR. As the presence of pseudogenes (numts) may be a concern in DNA barcoding (Song et al. 2008), sequences were screened for double peaks in the electropherograms as well as for the presence of indels and stop codons in an attempt to detect the potential presence of numts.
Distribution and diversity of haplotypes of H. halys
Statistical parsimony cladogram networks of the H. halys COI and Cyt B haplotypes were generated using the software package TCS v1.21 (Clement et al. 2000). For the construction of haplotype networks, sequences from specimens intercepted by border officials were included to provide a broader sampling of haplotypes from Asia and to provide qualitative information pertaining to potential pathways for entry. Measures of overall haplotype and nucleotide diversity for COI and Cyt B were generated using DnaSP v5.10.01 (Librado and Rozas 2009) and ARLEQUIN v3.1 (Excoffier et al. 2005).
Further, mitochondrial COI sequences from H. halys specimens were grouped into three broad categories based on the geographic area of collection: Asia, North America (Canada/USA), and Europe (Switzerland). Specimens intercepted by border officials were assigned to the geographic area from which a given shipment originated, as opposed to the geographic area in which they were intercepted. This provided a broader range of BMSB specimens from their Asian area of origin which would not have been attainable otherwise. Measures of genetic diversity were calculated for each of these three geographic regions using DnaSP v5.10.01 (Librado and Rozas 2009) and ARLEQUIN v3.1 (Excoffier et al. 2005), including: number of haplotypes, % of haplotypes unique to a given geographical area, haplotype diversity, and nucleotide diversity. Mean values for estimates of genetic variation in native and introduced populations were compared using an independent samples t test to determine whether a reduction in genetic variability was experienced upon introduction of BMSB to new locations.
Genetic structure among geographic regions was determined by AMOVA and pairwise FST analysis using ARLEQUIN v3.1 (Excoffier et al. 2005) in an effort to determine the strength of the associations within and between native and introduced populations. Haplotypes consistent between areas of introduction and area(s) of origin were used to speculate on the potential origin(s) of invading populations in Canada and Switzerland.
Results
DNA sequencing
Cytochrome Oxidase I: A total of 375 H. halys specimens (363 field-collected and 12 border interceptions) were obtained from North America, Europe, and Asia (Tables 1 and 2). All specimens yielded a 658-bp DNA sequence for the 5′ end of the COI gene. DNA sequence data and specimen collection information are on the Barcode of Life Database (BOLD), under the project Pentatomid Crop Pests (PENT). Corresponding Genbank accession numbers are as follows: KF273380–KF273402. None of the characteristic evidence of numts was present in the COI sequences analyzed, which is consistent with a larger study on the Hemiptera which included the Pentatomidae (Park et al. 2011).
28S: Although attempts were made to sequence all 375 H. halys specimens, amplification and sequencing success for the 28S gene was not successful for a number of specimens. As such, only 69 DNA sequences were obtained from the H. halys specimens processed. However, these specimens represented a geographically diverse group of samples collected or intercepted from Asia (n = 54; including intercepted specimens from Japan and Korea), North America (n = 6), and Europe (n = 9). No sequence variation was observed and despite the broad geographic origin of collected samples, all sequences for the 28S D2 region were identical (Genbank accession no. KF273403).
Cytochrome b: Sequences of 426 bp from the Cyt B gene were obtained for 232 of the 375 H. halys specimens. However, those specimens providing sequences represented samples collected or intercepted in Asia (n = 107), North America (n = 21), and Europe (n = 104). Genbank accession numbers for each unique haplotype are: KF273403–KF273412.
The COI gene provided the most complete sequence data for the specimens collected, and displayed the highest level of diversity in terms of haplotypes (see results below), and was thus used for more in-depth population genetic analysis related to recent establishment of H. halys in Canada and Switzerland.
Distribution and diversity of haplotypes of H. halys based on Cytochrome oxidase I
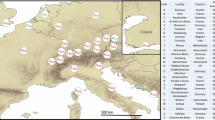
A total of 23 haplotypes (denoted H1–H23) were recovered from the samples, and consisted of 17 single nucleotide polymorphisms (Fig. 1; see supplementary Table 1 for detailed haplotype information on each sample). Five intermediate haplotypes were not represented in the H. halys collections. Among all samples collected, two haplotypes were dominant (H1 and H3) and were shared by ~85 % of the individuals collected. These two haplotypes were also the dominant haplotypes found in recently invaded areas in Canada (H1) and Switzerland (H3), respectively.
Statistical parsimony cladogram network representing the relationships among haplotypes for cytochrome oxidase 1 from H. halys collected in both native and introduced regions. Each circle is labeled by haplotype (H1 to H23) as well as the frequency (n) of the haplotype in the dataset; circle size is proportional to the frequency of each haplotype. Haplotypes that occur singly in the dataset were not labeled with a frequency (n) value. Differing shades in the pie diagrams indicate different geographic regions as indicated. Small, unlabeled circles indicate intermediate haplotypes not detected in our samples
The overall haplotype diversity was fairly high (0.628 ± 0.015 SD), and was significantly higher in native Asian populations of H. halys samples than in samples collected from recently invaded regions of Europe (t = 51.52, p < 0.0001) and North America (t = 63.49, p < 0.0001) (Table 3). Overall nucleotide diversity was relatively low (0.00145 ± 0.00008 SD), but significantly higher in native Asian samples than in either of the two recently invaded areas (Asia vs. Europe: t = 9.03, p < 0.0001; Asia vs. North America: t = 29.09, p < 0.0001). Results from the AMOVA showed that the overall genetic variation within populations accounted for 34.2 % of the variance, and was greater than the variation among populations (10.6 %). Pairwise FST values between native and invasive populations were significant (Asia vs. North America: FST = 0.054, p < 0.0001; Asia vs. Europe: FST = 0.526 p < 0.0001).
Twenty one of the 23 haplotypes were identified from the Asian samples, most of which were either H1 (67 %) or H3 (12.5 %). The remaining 19 haplotypes were represented by 1–3 individuals each (Table 4). Only individual specimens were obtained from Japan and Korea; however, they each represented unique haplotypes that were not found among samples collected elsewhere in Asia.
Samples from Switzerland contained representatives from three haplotypes, with the majority consistent with H3 (84.1 %), followed by H8 (15.4 %), and H9 (0.5 %) (Table 5). North American samples were predominantly the H1 haplotype (97 %), with individual representatives of two other haplotypes (H6 and H14) collected from private residences in ON, Canada. Specimens intercepted in Canada (n = 3) and Germany (n = 5) from shipments that originated in the USA were all haplotype H1. This was also consistent with lab-reared colonies originating from field-collected material in the USA. Only one specimen of Asian origin intercepted by border authorities in Canada was consistent with the dominant haplotype (H1) of the established population. The remaining interceptions originating from Asia represented haplotypes not detected in field-collected material in North America. Interestingly, the one interception of Asian origin that occurred geographically closest to the established breeding populations in Canada does not match any of the established populations in Canada. However, this haplotype is among the dominant haplotypes occurring in both Switzerland and China. Individuals intercepted from shipments originating in Korea and Japan did not match any of the recently established invasive populations.
All haplotypes in the North American samples were shared with the Asian samples from Hebei and Beijing provinces of China, and no unique haplotypes were identified in the invasive North American population. In contrast, only one of the haplotypes (H3) in the invaded area of Europe was consistent with specimens collected from both the Hebei and Beijing provinces. Although H3 was the dominant haplotype in Switzerland and was consistent with some of the Asian samples collected in the Hebei and Beijing provinces, it was not the dominant haplotype in these regions and thus the distribution and abundance of this haplotype in China is unclear. The remaining two haplotypes were unique to Switzerland and no matches were found among our Asian samples. Two of the haplotypes (H3 and H8) were found in all three Swiss Cantons sampled (Basel, Bern, and Zurich). The remaining unique haplotype (H9) was represented by a single specimen in the Zurich area.
Haplotypes of H. halys based on Cytochrome B
A total of 9 haplotypes representing 10 nucleotide polymorphisms were obtained from the 232 H. halys specimens analyzed, and two intermediate haplotypes were not represented among the samples (Fig. 2; see supplementary Table 1 for detailed haplotype information on each sample). Haplotype diversity (0.093 ± 0.026 SD) and nucleotide diversity (0.00028 ± 0.00010 SD) were low. Due to the relatively low diversity, this gene region did not provide adequate resolution for an in-depth analysis of invasion pathways of H. halys in areas where it has recently established. The dominant haplotype (95 % of all samples) was Cyt B haplotype H1, which included all specimens from the invasive populations from Canada/USA (n = 21) and Switzerland (n = 104) as well as most of the Asian samples (n = 96). The remaining eight haplotypes were represented by 1–2 individuals collected from Asia (n = 11). Individual samples falling within four of unique Cyt B haplotypes (Cyt B H2, H6, H7, and H9) also correspond to unique haplotypes detected using the COI gene (COI H2, H5, H17, and H23, respectively). All of these were from the Asian population of H. halys collected or intercepted from China (Beijing and Hebei provinces) and Japan. In contrast, Cyt B haplotype H1 encompassed all North American and Swiss samples and Asian samples within COI H1, H3, H4, H6–16, and H18–21. Additional Asian specimens within COI H1 corresponded to samples grouped within Cyt B H3, 4, 5, and 8 (see Table 6). Unfortunately COI haplotype H22 was not represented in the Cyt B sequence set as the sample failed to amplify and sequence. Otherwise, representatives of all other haplotypes were available for comparison. However, the variation in Cyt B haplotypes was not sufficient to address the origin and invasion pathways of BMSB in newly established populations and as such, further analysis on the Cyt B genetic data was not performed. However, Table 6 summarizes the COI and Cyt B haplotypes and Supplemental Table 1 lists each sample and the corresponding COI and Cyt B haplotypes to illustrate their distribution and highlight any inconsistencies between the two sets of data.
Statistical parsimony cladogram network representing the relationships among haplotypes for cytochrome b gene from H. halys collected in both native and introduced regions. Each circle is labeled by haplotype (H1 to H9) as well as the frequency (n) of the haplotype in the dataset; circle size is proportional to the frequency of each haplotype. Haplotypes that occur singly in the dataset were not labeled with a frequency (n) value. Differing shades in the pie diagrams indicate different geographic regions as indicated. Small, unlabeled circles indicate intermediate haplotypes not detected in our samples
Discussion
Many introduced species display severe genetic bottlenecks following introduction to a new environment, and have nonetheless become extremely successful colonizers (Sax and Brown 2000). The present study investigated the use of the mitochondrial COI and Cyt B genes in estimating the genetic variability and potential pathways of entry of H. halys in recently invaded areas. The COI gene demonstrated more variability than the Cyt B gene and was better suited for assessing the genetic variability and potential pathways of entry of invasive H. halys. The Cyt B gene showed both low haplotype and nucleotide diversity and was thus of limited use in the separation of the different populations. In fact, the vast majority of samples belonged to one haplotype (including all H. halys from recently invaded areas, as well as the majority of samples collected in China), and this particular Cyt B haplotype encompassed samples belonging to 18 different COI haplotypes. Interestingly, some discordance between the Cyt B and COI haplotypes was observed. As mentioned above, all of the invasive populations from North America (COI H1, H6, and H14 haplotypes) and Europe (COI H3, H8, and H9 haplotypes) fell within the Cyt B H1 haplotype. However, four individuals from the Chinese collections which displayed a COI H1 haplotype did not fall within the Cyt B H1 haplotype, but rather generated unique Cyt B haplotypes. This is interesting, as it may suggest the potential to further discriminate or subdivide the Chinese H. halys population using the Cyt B gene in combination with the COI gene. However, the utility and validity of this approach would need to be explored using a large number of samples across a broad geographic range in the area of origin of H. halys.
Our COI data show that recently established populations of H. halys have reduced genetic variability in comparison to native Asian populations. Nucleotide and haplotype diversity in the two recently established populations was significantly lower than that in the area of origin (three haplotypes in invaded regions vs. 21 in Asia), presumably due to founder effects during colonization of North America and Switzerland.
Three haplotypes were discovered among the North American samples, and all three of these haplotypes were shared with native Chinese samples from Hebei and Beijing provinces. However, a single haplotype dominated the North American samples, representing 96 % of the specimens from Canada and the USA. Established breeding populations of H. halys in Canada belong to one single haplotype. The occurrence of two additional, singly occurring haplotypes from residential homes in Canada is likely not representative of the field-collected population, but it demonstrates the value of including homeowner finds (versus only field-collected specimens) in haplotype diversity studies for H. halys. This may be particularly important in the early stages of the establishment of an invasive species in a new area, as it allows the detection of additional and/or rare haplotypes. The occurrence of such events suggests the potential for additional haplotypes to become established from multiple, independent introduction events from the native range of H. halys, and may suggest routes of entry that serve to increase the genetic diversity of invasive BMSB populations over time.
Historical interception records for the arrival of H. halys in Canada indicate that this species could have arrived from Asia in the early 1990’s. However, most of these early interceptions occurred on Canada’s west coast (British Columbia), where no homeowner finds or established populations have been reported to date. Only one of the intercepted Asian specimens collected by phytosanitary officers in Canada was consistent with the dominant, established haplotype in Canada. Although the shipment origin was listed as China, the exact geographic location within the country remains unknown; however, it matches the dominant haplotypes from the Beijing and Hebei Provinces in China so we suspect that it originated from this region, but further evidence is needed to confirm this observation. This single specimen was intercepted in Vancouver, British Columbia (2008) which is far removed (>4,000 km) from the location where H. halys has become established. We, therefore, assume that this was not the original source population responsible for the establishment of BMSB in Canada. Confirmed homeowner reports of H. halys in eastern Canada (southern Ontario) have been consistent and growing since 2010, indicating the likely arrival and establishment around that time. This coincides with the extreme population explosion of H. halys in the North Eastern USA, as well as with the increase in H. halys interceptions in eastern Canada originating from the USA. The haplotype identity and timing of H. halys population establishment in Canada are consistent with the movement or expansion of US populations due to human activity (trade, travel, etc.). It is thus more likely that established populations in southern Ontario are derived from movement of the population in the eastern USA into Canada, with the likely initial source population originating from either Beijing or Hebei Provinces in China, given the dominance of Haplotype H1 from these regions. This supports recently collected sequence data for the COII gene, which also points to China as the likely source population of H. halys in North America (Xu et al. 2013). However, it is possible that other regions in China also have this haplotype, and as such further investigation is needed to determine the distribution and prevalence of this haplotype from a broader geographic range in China, particularly given the propensity for movement of this pest by human-mediated activity.
Historical records for the occurrence of H. halys interceptions in Switzerland were not available, and thus the pathways for entry are unclear. Our collections in 2012 showed that three haplotypes were also represented in the Swiss population. Only one haplotype was shared between the Swiss population and the Asian population, and no haplotypes were shared between the Swiss and Canadian populations. In contrast to the Canadian occurrence, two of these haplotypes occur frequently (representing 84 and 15.5 % of the Swiss samples) in the established breeding populations and homeowner finds in at least three Cantons. This suggests the possibility of multiple invasion events from different locations, or a single introduction event with a larger number of genetically distinct individuals. A third haplotype was also detected in the Zurich population, but it occurred singly and does not appear to be representative of the established field population. As mentioned above, occurrence of additional, rare haplotypes may indicate further introduction events and new sources of diversity for genetically restricted founding populations. As no unique haploytpes were found outside the Canton Zurich, our data suggest that the movement of the founding population in Zurich is responsible for seeding the population in other Cantons within the country, as opposed to additional introductions from Asia or North America. It is highly likely that following the introduction and establishment of these haplotypes in the Zurich area, populations expanded to additional Cantons through human activity and movement of goods within the country in a “stepping stone” expansion of the original introduction(s).
Interestingly, none of the haplotypes were shared between the North American and Swiss samples, which provides a clear indication that the two invasions occurred separately, and from different source populations (i.e., no admixture in any of the introduced populations). This provides conclusive evidence that the established population of H. halys in Switzerland did not occur due to trade between the USA and Europe, despite previous interceptions of H. halys in Europe from shipments originating in Pennsylvania, USA (Freers 2012). Interestingly, one of the samples intercepted in Canada in 2001 (Strathroy, Ontario) matched the dominant haplotype (H3) in Switzerland, which is shared with the Chinese population (Beijing and Hebei Provinces). Unfortunately, the exact area of origin for this shipment from China was not documented, and it provides no indication as to the location where this individual came from. Thus, the area of origin of the Swiss population(s) is still unresolved, as two of the three haplotypes were unique to Switzerland and were not in common with the Asian samples collected. Further, sampling from across the broad geographic range of H. halys in Asia, including Korea, Japan, and Taiwan (Zhu et al. 2012) would clarify the origin of the two haplotypes that are unique to the established populations in Switzerland. This would also be advantageous for biological control, because explorations for natural enemies of H. halys could then be focused on the native range of the best genetically matched H. halys populations, assuming that natural enemies in the geographic centre of origin are better adapted to the pest (Hoelmer and Kirk 2005).
Our Asian collections were fairly limited in geographic scope, and as such more extensive sampling throughout Asia would likely result in a haplotype match and a better understanding of the route and mode of entry of the established haplotypes in Switzerland. As such, we assume that the diversity in Asia is actually much higher than that reflected in our samples. Although the present data suggest that specimens from the Hebei and/or Beijing Provinces established in North America, we assume that additional haplotypes from within China would permit a more conclusive identification of the source population occurring in North America. It is important to note that although our sampling efforts were not widespread across Asia, large numbers of individuals were collected from the Hebei and Beijing Provinces (108 individuals). As such, we are confident that the haplotype diversity and distribution in this particular region have been adequately captured in comparison to studies which cover a broader geographic range with a limited number of specimens at each locality (e.g., 1–5 specimens per region). Based on climate and distribution data, Zhu et al. (2012) predicted that the source of the invasive populations in North America and Europe would be either western South Korea or northern Japan. However, our data are in disagreement with their model (at least in terms of the North American samples), and suggest that the source of H. halys in North America is most likely from China. Further sampling in Asia should help pinpoint the source(s) of H. halys in Europe and could be used to re-evaluate the climate model proposed by Zhu et al. (2012).
It will be important to continue to monitor the genetic diversity of these recently introduced populations to determine whether variability will increase over time as additional colonization/introduction events take place. Hybridization of individuals from genetically distinct populations (e.g., from China and Japan) may lead to an increased fitness (e.g., higher fecundity) of H. halys populations in the invaded area (Reed and Frankham 2003; Szucs et al. 2012) and admixed populations may be better adapted to new environments, which may further compound the current problems associated with the invasive pest. More targeted trapping and monitoring programs in recently invaded areas could provide additional information on movement and establishment success of different haplotypes. Ultimately, identifying the origin of invasive H. halys populations may help to detect unknown pathways and stop the potential influx of additional haplotypes contributing to the genetic diversity of this pest.
References
Aldrich JR, Khrimian A, Chen X, Camp MJ (2009) Semiochemically based monitoring of the invasion of the brown marmorated stink bug and unexpected attraction of the native green stink bug (Heteroptera: Pentatomidae) in Maryland. Fla Entomol 92:483–491
Arnold K (2009) Halyomorpha halys (Stål, 1855), eine für die europäische Fauna neu nachgewiesene Wanzenart (Insecta: Heteroptera: Pentatomidae: Cappaeini). Mitt Thüringer Entomol 16:19
Auger-Rozenberg MA, Boivin T, Magnoux E, Courtin C, Roques A, Kerdelhué C (2012) Inferences on population history of a seed chalcid wasp: invasion success despite a severe founder effect from an unexpected source population. Mol Ecol 21:6086–6103
Australian Department of Agriculture, Fisheries and Forestry (2012a) Operational science program bulletin December 2011/January 2012. http://www.daff.gov.au/__data/assets/pdf_file/0004/2160481/PDF_version_Jan_2012_OSP_Bulletin.pdf. Accessed 21 Jan 2013
Australian Department of Agriculture, Fisheries and Forestry (2012b) Operational science program bulletin February 2012. http://www.daff.gov.au/__data/assets/pdf_file/0007/2160817/osp-bulletin-feb12.pdf. Accessed 21 Jan 2013
Avise JC (1995) Mitochondrial DNA polymorphism and a connection between genetics and demography of relevance to conservation. Conserv Biol 9:686–690
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522
Barr NB (2009) Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J Econ Entomol 102:401–411
Blanchet S (2012) The use of molecular tools in invasion biology: an emphasis on freshwater ecosystems. Fish Manag Ecol 19:120–132
Callot H, Brua C (2013) Halyomorpha halys (Stål, 1855), la Punaise diabolique, nouvelle espèce pour la faune de France (Heteroptera Pentatomidae). L’Entomologiste 69:69–71
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Corin SE, Lester PJ, Abbott KL, Ritchie PA (2007) Inferring historical introduction pathways with mitochondrial DNA: the case of introduced Argentine ants (Linepithema humile) into New Zealand. Divers Distrib 13:510–518
Cuituan Z, Daluan L, Haifeng S, Guoliang X (1993) A study on the biological characteristics of Halyomorpha picus and Erthesina fullo. For Res 3:5
deLeón JH, Sétamou M, Gastaminza GA, Buenahora J, Cáceres S, Yamamoto PT, Bouvet JP, Logarzo GA (2011) Two separate introductions of Asian citrus psyllid populations found in the American continents. Ann Entomol Soc Am 104:1392–1398
Duthie C (2012) Risk analysis of Halyomorpha halys (Brown Marmorated Stink Bug) on all pathways. Ministry for Primary Industries, Wellington, p 57
Estoup A, Guillemaud T (2010) Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol 19:4113–4130
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fogain R, Graff S (2011) First records of the invasive pest, Halyomorpha halys (Hemiptera: Pentatomidae), in Ontario and Quebec. J Entomol Soc Ont 142:45–48
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Franks SJ, Pratt PD, Tsutsui ND (2011) The genetic consequences of a demographic bottleneck in an introduced biological control insect. Conserv Genet 12:201–211
Freers A (2012) Blinde Passagiere: Stinkwanzen, Marmorierte Baumwanze–Halyomorpha halys. https://ssl.bremen.de/lmtvet/sixcms/media.php/13/Blinde_Passagiere_Stinkwanze_3_2012.pdf. Accessed 11 March 2013
Funayama K (2003) Outbreak and control of stink bugs in apple orchard. Jpn Agric Technol 47:35–39
Grapputo A, Boman S, Lindstroem L, Lyytinen A, Mappes J (2005) The voyage of an invasive species across continents: genetic diversity of North American and European Colorado potato beetle populations. Mol Ecol 14:4207–4219
Hasegawa H, Umeya K (1974) Outbreak of stink bugs in fruit trees. Plant Prot 28:279–286
Haye T, Wyniger D (2013) Current distribution of Halyomorpha halys in Europe. http://www.halyomorphahalys.com. Accessed April 2013
He YB, Wan XW, Liu YH, Sun GM, Zhan RL (2012) Mitochondrial COI from Dysmicoccus brevipes (Hemiptera: Pseudococcidae) suggests cryptic lineage and pinpoints the source of the introduction to China. Fla Entomol 95:183–191
Heckmann R (2012) Erster Nachweis von Halyomorpha halys (STÅL, 1855) (Heteroptera: Pentatomidae) für Deutschland. Heteropteron 36:17–18
Hoebeke ER, Carter ME (2003) Halyomorpha halys (Stål) (Heteroptera: Pentatomidae): a polyphagous plant pest from Asia newly detected in North America. P Entomol Soc Wash 105:225–237
Hoelmer KA, Kirk AA (2005) Selecting arthropod biological control agents against arthropod pests: can the science be improved to decrease the risk of releasing ineffective agents? Biol Control 34:255–264
Hulme PE (2009) Trade, transport and trouble: managing invasive species pathways in an era of globalization. J Appl Ecol 46:10–18
Hulme PE, Bacher S, Kenis M, Klotz S, Kühn I, Minchin D, Nentwig W, Olenin S, Panov V, Pergl J (2008) Grasping at the routes of biological invasions: a framework for integrating pathways into policy. J Appl Ecol 45:403–414
Kaňuch P, Berggren Å, Cassel-Lundhagen A (2012) Colonization history of Metrioptera roeselii in northern Europe indicates human-mediated dispersal. J Biogeogr 40:977–987
Kavar T, Pavlovcic P, Susnik S, Meglic V, Virant-Doberlet M (2006) Genetic differentiation of geographically separated populations of the southern green stink bug Nezara viridula (Hemiptera: Pentatomidae). Bull Entomol Res 96:117–128
Leskey TC, Short BD, Butler BR, Wright SE (2012a) Impact of the invasive brown marmorated stink bug, Halyomorpha halys (Stål), in Mid-Atlantic tree fruit orchards in the United States: case studies of commercial management. Psyche 2012:1–14
Leskey TC, Wright SE, Short BD, Khrimian A (2012b) Development of behaviorally based monitoring tools for the brown marmorated stink bug, Halyomorpha halys (Stål)(Heteroptera: Pentatomidae) in commercial tree fruit orchards. J Entomol Sci 47:76–85
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Mayr E (1954) Change of genetic environment and evolution. In: Huxley J, Hardy A, Ford E (eds) Evolution as a process. Allen and Unwin, London, pp 157–180
Miller N, Estoup A, Toepfer S, Bourguet D, Lapchin L, Derridj S, Kim KS, Reynaud P, Furlan L, Guillemaud T (2005) Multiple transatlantic introductions of the western corn rootworm. Science 310:992
Mueller G, Luescher IL, Schmidt M (2011) New data on the incidence of household arthropod pests and new invasive pests in Zurich (Switzerland). In: Proceedings of the seventh international conference on urban pests: 99–104
Muraji M, Kawasaki K, Shimizu T (2000) Phylogenetic utility of nucleotide sequences of mitochondrial 16S ribosomal RNA and cytochrome b genes in anthocorid bugs (Hemiptera: Anthocoridae). Appl Entomol Zool 35:293–300
Nielsen AL (2011) Invasion biology and seasonality of BMSB in Pennsylvania and New Jersey (presentation 0020). In: Proceedings of the 59th annual meeting of the entomological society of America, 13–16 Nov 2011
Nielsen AL, Hamilton GC (2009a) Life history of the invasive species Halyomorpha halys (Hemiptera: Pentatomidae) in Northeastern United States. Ann Entomol Soc Am 102:608–616
Nielsen AL, Hamilton GC (2009b) Seasonal occurrence and impact of Halyomorpha halys (Hemiptera: Pentatomidae) in tree fruit. J Econ Entomol 102:1133–1140
Nielsen AL, Hamilton GC, Shearer PW (2011) Seasonal phenology and monitoring of the non-native Halyomorpha halys(Stål) (Hemiptera: Pentatomidae) in soybean. Environ Entomol 40:231–238
Park D-S, Foottit R, Maw E, Hebert PDN (2011) Barcoding bugs: DNA-based identification of the true bugs (Insecta: Hemiptera: Heteroptera). PloS One 6:e18749
Piel F, Gilbert M, De Cannière C, Grégoire JC (2008) Coniferous round wood imports from Russia and Baltic countries to Belgium. A pathway analysis for assessing risks of exotic pest insect introductions. Divers Distrib 14:318–328
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Sax DF, Brown JH (2000) The paradox of invasion. Global Ecol Biogeogr 9:363–371
Smith KF, Stefaniak L, Saito Y, Gemmill CEC, Cary SC, Fidler AE (2012a) Increased inter-colony fusion rates are associated with reduced COI haplotype diversity in an invasive colonial ascidian Didemnum vexillum. PLoS One 7:e30473
Smith KF, Thia J, Gemmill CEC, Cary SC, Fidler AE (2012b) Barcoding of the cytochrome oxidase I (COI) indicates a recent introduction of Ciona savignyi into New Zealand and provides a rapid method for Ciona species discrimination. Aquat Invasions 7:305–313
Song H, Buhay JE, Whiting MF, Crandall KA (2008) Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA 105:13486–13491
Stephens JD, Santos SR, Folkerts DR (2011) Genetic Differentiation, structure, and a transition zone among populations of the pitcher plant moth Exyra semicrocea: implications for conservation. PLoS One 6:e22658
Szűcs M, Eigenbrode SD, Schwarzländer M, Schaffner U (2012) Hybrid vigor in the biological control agent, Longitarsus jacobaeae. Evol Appl 5:489–497
Teske PR, Rius M, McQuaid CD, Styan CA, Piggott MP, Benhissoune S, Fuentes-Grünewald C, Walls K, Page M, Attard CRM (2011) “Nested” cryptic diversity in a widespread marine ecosystem engineer: a challenge for detecting biological invasions. BMC Evol Biol 11:1–13
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walker K (2009) Brown marmorated stink bug (Halyomorpha halys). PaDIL. http://www.padil.gov.au. Accessed 21 Jan 2013
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506
Watanabe M, Arakawa R, Shinagawa Y, Okazawa T (1994) Anti-invading methods against the brown marmorated stink bug, Halyomorpha mista, in houses. Jpn J Sanit Zool 45:311–317
Wermelinger B, Wyniger D, Forster B (2008) First records of an invasive bug in Europe: Halyomorpha halys Stal (Heteroptera: Pentatomidae), a new pest on woody ornamentals and fruit trees? Mitt Schweiz Entomol Ges 81:1–8
Wyniger D, Kment P (2010) Key for the separation of Halyomorpha halys (Stål) from similar-appearing pentatomids (Insecta: Heteroptera: Pentatomidae) occurring in Central Europe, with new Swiss records. Mitt Schweiz Entomol Ges 83:261–270
Xu J, Fonseca DM, Hamilton GC, Hoelmer KA, Nielsen AL (2013) Tracing the origin of US brown marmorated stink bugs, Halyomorpha halys. Biol Invasions (in press)
Zhu G, Bu W, Gao Y, Liu G (2012) Potential geographic distribution of brown marmorated stink bug invasion (Halyomorpha halys). PLoS One 7:e31246
Acknowledgments
The authors are grateful to all Swiss homeowners who kindly provided us with specimens, particularly A. Knup, A. Six, R. Burtscher, W. and T. Rüesch, E. Roschet, and K. Häberlin. We would like to thank the team of the MoA-CABI Joint Lab for Biosafety for their assistance and Dr. Y. Lu for kindly giving us access to his light traps at the Langfang Experimental Station. The authors would like to thank Allison Bruin for technical assistance, and the Canadian Food Inspection Agency and the Canadian National Collection, particularly O. Lonsdale, for providing intercepted specimens in shipments and cargo.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Traugott.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gariepy, T.D., Haye, T., Fraser, H. et al. Occurrence, genetic diversity, and potential pathways of entry of Halyomorpha halys in newly invaded areas of Canada and Switzerland. J Pest Sci 87, 17–28 (2014). https://doi.org/10.1007/s10340-013-0529-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-013-0529-3