Abstract
Generalist arthropod predators are potential drivers of population dynamics in a wide variety of ecosystems but their feeding habits are often difficult to reveal as they are small, mobile, and live among dense vegetation or in soils. DNA-based gut-content analysis is a powerful tool that enables studies on arthropod predator–prey interactions. We studied lycosid spiders (Pardosa spp.) in agroecosystems to see if they consumed cereal aphids (Rhopalosiphum padi) and Collembolans at random, i.e., in proportion to their abundance in the field. We also tested if consumption of the target prey items was affected by the presence of alternative food. Spiders were captured in farmers’ fields and their gut-contents screened by PCR with R. padi and Collembola primers. On all sampling occasions, concurrent assessments of total prey availability were carried out. Spider predation rates on R. padi always exceeded 50 %. Spiders also tested positive for Collembola but to a lower and more varying degree. In general, Pardosa did not consume R. padi and Collembolans in relation to their abundance in the field. Aphid predation was much higher than expected whereas consumption of Collembolans was considerably lower. The presence of alternative prey influenced consumption of the aphid. It was concluded that prey consumption by Pardosa spiders generally cannot be assumed to simply mirror prey availability. The spatial distribution of the target prey needs to be considered as well as the abundance, composition, and nutritional content of potential alternative food items.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Detailed knowledge about the foraging behavior of predators is fundamental in food web ecology and necessary to make correct assumptions about the biological control potential of predators (Beckerman et al. 2006). In many cases, however, information about specific predator–prey interactions and how such pathways are affected by the presence of co-occurring species is limited (Schmitz 2007). In the case of arthropod predators, the lack of knowledge largely depends on difficulties associated with direct observation of small, mobile organisms that live under dense vegetation or below the soil surface. In addition, many arthropod predators are generalists and may include a wide range of prey items in their diet, which complicates studies on their feeding behavior in their natural habitats even further (Symondson et al. 2002).
One way to obtain specific information about the diet of arthropod predators, under a minimal level of disturbance, is to collect predators in their natural environment and analyze prey remains that are present in their gut-contents. Currently, detection of prey DNA using PCR-based techniques is the most commonly used approach (reviewed by Sheppard and Harwood 2005; King et al. 2008). If consumption data derived from such analyses are combined with assessments of prey availability new insights about a predator’s feeding behavior or hunting strategy can be obtained.
In agricultural habitats, generalist predators such as ground beetles and spiders feed on crop pests (Symondson et al. 2002; Kuusk et al. 2008; Tahir and Butt 2009; Öberg et al. 2011). Under certain conditions, they have the potential to maintain pest populations below economic thresholds and thus provide an important ecosystem service to the agricultural sector acting as biological control agents (Chiverton 1986; Östman et al. 2001, 2003). In addition to specific pests, generalist predators may consume alternative food items including other herbivores, decomposers such as earthworms and Collembola (Juen and Traugott 2007; King et al. 2010; Kuusk and Ekbom 2010), Dipterans (Harwood et al. 2007), and other natural enemies (Harwood et al. 2009). By comparing rates of predation derived from molecular gut-content assays with the observed proportion of the target prey in the field, the hypothesis that generalist predators are capturing prey at random can be tested. Such tests have been carried out but are so far restricted to only a few combinations of predators and prey; linyphiid spiders and Collembolans by Agusti et al. (2003), linyphiid spiders and aphids, dipterans, Collembolans (Harwood et al. 2004, 2007), carabid beetles and earthworms by King et al. (2010), coccinellid larvae and dipterans (Moser et al. 2011), carabid beetles and slugs (Hatteland et al. 2011). Therefore, there is a clear need for more data on other taxa.
Previously, we studied the foraging behavior of generalist spider predators from the genus Pardosa (Araneae: Lycosidae) in farmer’s fields in central Sweden by combining DNA-based gut-content analysis and concurrent monitoring of prey availability (Kuusk et al. 2008; Kuusk and Ekbom 2010). Pardosa spiders are common ground-living predators in European agricultural habitats (Samu and Szinetár 2002; Schmidt et al. 2005; Öberg et al. 2007), they have good eyesight and some species are viewed as active hunters that localize their prey by visual cues (Foelix 1996). Others, which feed on highly mobile prey, hunt with a “sit-and-wait strategy,” i.e., they lie in ambush and wait for a prey to come close enough to be overpowered (Edgar 1969). Samu et al. (2003) pointed out that sit-and-wait and actively foraging strategies represent two ends of a continuum and introduced another hunting category. Based on visual observations in alfalfa Pardosa agrestis (Westring) was classified as a “sit-and-move forager” that indeed spends most of the time sitting still but will frequently change their waiting positions.
We have focused on Pardosa’s consumption of the bird-cherry oat aphid, Rhopalosiphum padi (L.) (Homoptera: Aphididae) and springtails (Arthropoda: Collembola). The aphid is a major pest in spring-sown cereals in Northern Europe (Wiktelius and Ekbom 1985; reviewed by Leather et al. 1989) whereas Collembolans are primarily detritivores that represent a substantial source of alternative prey to generalist arthropod predators worldwide (Hopkin 1997). Our previous results were derived from DNA-based gut-content analyses of spiders collected in fields where R. padi densities were consistently very low. We demonstrated that Pardosa spiders did indeed feed on R. padi at such low pest densities, which is necessary if they are to suppress the aphid population below economic thresholds (Chiverton 1986; Östman et al. 2001). We also showed that Pardosa use Collembolans as alternative food and that there was a negative association between the probability of detecting aphid DNA in individual spiders and Collembola availability. This result was puzzling as we did not observe a positive relationship between Collembola consumption and Collembola availability. We suggested that the spiders changed their hunting strategy when ratios of sedentary and mobile prey changed. When the quick, jumping Collembolans were numerous, Pardosa hunted more passively with a “sit-and-wait” strategy and consequently did not encounter so many R. padi which are sedentary on plant stem bases when present in low densities. In contrast, when Collembolans were less abundant, Pardosa had to forage more actively to encounter more prey and consequently “ran into” more aphids.
The overall purpose of this study was to extend our knowledge about the foraging behavior and hunting strategy of Pardosa spiders. In line with previous studies, PCR-based gut-content analysis was used to screen field-collected predators for R. padi and Collembola consumption and at each sampling occasions concurrent assessments of prey availability were carried out. In contrast to previous studies, the collected spiders were exposed to both low and very high numbers of R. padi as well as varying abundances of different alternative prey.
The specific aims included to find out (1) if the probability of detecting R. padi and Collembolans in individual spiders was related to prey availability and (2) if R. padi predation was affected by the presence of alternative prey taxa.
Materials and methods
Collection of spiders in spring-sown cereals
Adult Pardosa spiders of both sexes, including P. agrestis, P. palustris (L.), P. prativaga (L. Koch), P. fulviseps (Collet), and P. amentata (Clerk), were randomly collected by hand from three organic farms in fields of spring-sown cereals around Uppsala, Sweden (59°51′N, 17°41′E). The shortest distance between farms was 19 km and the longest distance was 46 km. Each field was sampled on two occasions (Table 1); the first at the start of stem elongation of the crop and the second during booting. The collection area within each field measured 60 × 40 m2 and was located 5–10 m from the field edge. All collections were carried out between 0900 and 1400 h and captured predators were immediately transferred to individual Eppendorf tubes and frozen using dry ice. Spiders were stored at −70 °C prior to DNA extraction and PCR analysis described below. On one occasion during the storage period the predators were briefly handled to identify species and sexual category (males, females carrying an eggsac, females without an eggsac).
Assessments of natural prey abundances
An effective method of monitoring the availability of prey is the use of mini-sticky traps; Harwood et al. (2001, 2004) used such traps for studying the availability of prey to linyphiid spiders in agricultural habitats. We used a similar method to assess prey availability modified for wolf spiders, 12 mini-sticky traps (10 × 5 cm2) were randomly attached to the ground in each collection area. The catches represented a cumulative record of prey over 24 h starting in the afternoon 1 day prior to the collection of predators and ending when the capture session terminated (for more details about the traps see Kuusk and Ekbom 2010). Captured arthropods were identified to the following taxa: Aphidoidea, Collembola, Diptera, Cicadoidea, Thysanoptera, Hymenoptera, Coleoptera, Araneae, and others. Mean prey abundances per trap and SE were calculated for each sampling occasion as well as the proportion of each taxa of the total prey community. Apart from assessing aphid densities on the ground with mini-sticky traps we also estimated the mean number of aphids per plant at each sampling occasion by randomly selecting 50–100 plants in the collection area; plants were gently removed from the soil so that both above and below ground aphids could be counted (Kuusk et al. 2008). Mini-sticky traps will most likely catch aphids that have fallen off plants and these aphids will be available to the wolf spiders. Bird cherry-oat aphids will often occur at the base of the plants (Wiktelius 1987) and also be available to the wolf spiders. Therefore, the two measurements were finally combined to represent the total availability of aphids per 50 cm2 (the surface covered by one trap) (Table 1). From the plant counts means per shoot were multiplied by 1.25 to convert them to means per 50 cm2 area (we used a plant density of 250 plants per m2; this is based on recommendations to sow to a density of 350 plants per m2 and experience of an approximate establishment rate of 75 %) and then added to the means from the mini-sticky traps. The combined measurement of R. padi availability was then used in the chi-square goodness-of-fit test and the logistic regression analyses described below.
DNA extraction and PCR conditions
DNA was extracted from whole spiders using Qiagen’s DNeasy® Blood and Tissue Kit following the manufacturer’s instructions for animal tissue. To ascertain that the DNA extractions were successful, the quality of all samples was checked using the universal invertebrate primers LR-J-12887 and LR-N-13398 (Simon et al. 1994). Two extracts did not yield any PCR product and were excluded from the study. Subsequently, the samples were screened for R. padi consumption using primers developed by Chen et al. (2000) and a protocol described in Kuusk et al. (2008). Collembola consumption was tracked using group-specific Collembola primers and the associated protocol developed by Kuusk and Agustí (2008). In each run, diluted prey DNA and water were used as positive and negative controls, respectively. As additional controls we used DNA extracts from starved spiders that had been fed with either a single R. padi or a Collembola. PCR products were separated by electrophoresis in agarose gels stained with ethidium bromide and visualized under UV light. A sample was scored as positive when a band of approximately 331 bp (R. padi product) or 272 bp (Collembola product) appeared on the gel. All negative samples were run a second time and if a band appeared on the gel they were scored positive.
Detection times can vary depending on the combination of predator and prey and it may be necessary to adjust field predation rates (Greenstone et al. 2010; Gagnon et al. 2011). Previous feeding trials with Pardosa spiders, R. padi, and Collembola, conducted at the same temperatures and using the same primers as those used here, showed that aphid detection success over time for 50 % of the predators was 3.7 h (Kuusk et al. 2008) and no springtail DNA could be detected after 24 h (Kuusk and Ekbom 2010). Although we cannot say that there is no significant difference in detection times for the two preys, the feeding trials indicate that the detection times are fairly close and adjustments would not change our conclusions.
Statistical analyses
To test the hypothesis that Pardosa spiders feed at random without any preference for a particular prey, the proportion of spiders that tested positive by PCR was compared with the observed proportions of R. padi and Collembolans in the field using a chi-square goodness-of-fit test. For each sampling occasion, the spiders were divided into four categories: (1) individuals positive for both aphid and Collembola DNA, (2) individuals positive for aphid but not Collembola DNA, (3) individuals positive for Collembola but not aphid DNA, and (4) individuals negative for both preys. We assumed for the purposes of the test that the probability that a spider would contain aphid or Collembola DNA was directly proportional to the observed proportion of prey at each sampling occasion and that the events “eating aphid” and “eating Collembola” were independent.
To investigate which factors may have affected R. padi and Collembola consumption, the PCR data were analyzed using logistic regression (Hosmer and Lemeshow 2000); the SAS LOGISTIC procedure with the model binary logit. To correct for possible overdispersion we used the “scale = deviance” option in SAS. The presence or the absence of R. padi and Collembola DNA were the dependent variables in two separate analyses. As P. prativaga, P. fulviseps, and P. amentata each represented <5 % of the total number of collected spiders, data for these species were not included in the analysis. We tested if consumption of aphids was affected by farm, Pardosa species (P. agrestis and P. palustris), sexual category (males, females with eggsac, females without eggsac), or prey availability in multiple logistic regressions (Table 2). The abundance of R. padi and the sum of all alternative prey (Collembola, Diptera, and other prey; Table 1) were used for prey availability in the analysis of R. padi consumption. The abundance of Collembola and the sum of all alternative prey (R. padi, Diptera and other prey; Table 1) were used in the analysis of Collembola consumption.
Results
Of all analyzed spiders (n = 231), 71 % tested positive for the presence of R. padi DNA and 19 % for the presence of Collembola DNA. We analyzed 28–51 spiders per collection occasion and the proportions that tested positive per field for R. padi and Collembola DNA varied between 51–87 % and 3–62 %, respectively (Table 1). Between 3 and 50 % of the tested individuals scored positive for both preys (Table 1).
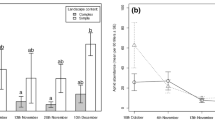
The availability of R. padi varied considerably between collection occasions, ranging from approximately 0.1–41 individuals per plant and 0.25–11 individuals per trap (Table 1). The total amount of available R. padi per sampling occasion (aphids per plant and trap combined) varied between approximately 0.5 and 60 individuals per 50 cm2. Mean abundances of Collembola, Diptera, and the pooled number of other potential prey varied between approximately 5 and 110, 3 and 50, and 1 and 3 per sampling occasion, respectively (Table 1). The proportions of R. padi, Collembola and other alternative prey of the total prey community are shown in Fig. 1.
Availability of R. padi (black bars), Collembola (white bars), and other alternative prey (gray bars) in three fields of spring-sown cereals at organic Farms A–C in central Sweden shown as percentages of the total prey community. Black squares and open circles show the proportion spiders tested positive for R. padi and Collembola DNA-remains, respectively, using PCR-based gut-content analysis
The most common Pardosa species was P. agrestis (78 %), followed by P. palustris (16 %), P. prativaga (4 %), and others (2 %). Of the two most common species, 69 and 82 % tested positive for aphid predation whereas 13 and 39 % contained Collembola DNA. Of all collected spiders, 30 % were males, 56 % females carrying an eggsac, and 14 % were females without an eggsac. In each sexual category, 68, 71, and 76 % tested positive for R. padi DNA, respectively. The corresponding data for Collembolans were 29, 18, and 3 %.
For five out of six collection occasions, the chi-square goodness-of-fit test clearly demonstrated that Pardosa spiders did not predate on R. padi and Collembolans in proportion to their abundances in the fields (Farm A June 16: χ 23 = 1030, P < .001; Farm A June 26: χ 23 = 833, P < .001; Farm B June 19: χ 23 = 120, P < .001; Farm B June 29: χ 23 = 49, P < .001; Farm C June 21: χ 23 = 276, P < .001). Aphid predation was much higher than what would be expected whereas the consumption of Collembolans was considerably lower (Fig. 1). For both prey types, the exception was Farm C June 30 (Fig. 1). On this collection occasion, the proportion of spiders that tested positive for R. padi and Collembola consumption corresponded to the availability of each prey (χ 23 = 0.4, P > 0.9).
In the logistic regression analysis there was some overdispersion in the data and the covariance matrix for each model was multiplied by the heterogeneity factor (R. padi analysis = 1.44, Collembola analysis = 1.10). The models were significant and the fit was adequate (Table 2). The results showed that neither Pardosa species nor sexual category influenced the probability of detecting R. padi or Collembola DNA in individual spiders; but the probability was significantly affected by the farm where samples were collected (Table 2).
Considering the effect of the variation in prey abundance for the probability of detecting R. padi DNA, we found that there was no association between R. padi consumption and R. padi availability (Table 2). In contrast, alternative prey densities appeared to influence R. padi consumption (P = 0.04). The relationship with alternative prey availability was positive (Table 2).
The logistic regression analysis of the probability of detecting Collembola DNA and prey abundance showed that there was a weak, positive relationship between Collembola availability and Collembola consumption (P = 0.065, Table 2). There was, however, no influence of alternative prey (Table 2) on the probability of detecting Collembola DNA in spiders.
Discussion
This work has generated new information about the feeding habits and foraging behavior of lycosid spiders from the genus Pardosa. We tested if consumption of two types of prey, an aphid pest (R. padi) and detritivorous Collembolans corresponded to the ratios at which the food items were present in farmers’ fields. In general, far more spiders contained R. padi DNA than expected while the reverse was true for Collembolans (Fig. 1). In total, 70 % of the analyzed specimens contained R. padi DNA and predation rates exceeded 50 % at all sampling occasions (Fig. 1.). The results suggest that Pardosa spiders readily ate R. padi and that this aphid formed a substantial part of the predator’s diet. The same trend was found in a previous study when aphid numbers were very low, far more spiders than expected tested positive for aphid DNA (Kuusk et al. 2008).
From one point of view this is surprising as laboratory studies have shown that a pure diet of R. padi is detrimental for cereal spiders with respect to various fitness parameters (Toft 1995; Oelbermann and Scheu 2002). In addition, it has been demonstrated that naïve Pardosa only have to consume a few R. padi individuals before developing an aversion against them (Toft 1997). On the other hand, ingestion of small amounts of R. padi together with other prey has proven superior to diets without any R. padi supplements indicating that the aphid contains essential nutrients (Toft 1995). Apart from nutritional aspects, another explanation to the disproportionally high predation rates of the aphid is that R. padi should be a fairly easy prey for the spiders to overpower. Aphids both on plants and on the ground were almost exclusively wingless individuals without any morphological characters that would impede a several times larger and faster predator from attacking.
Aphid predation rates for two subfamilies of Linyphiidae collected in winter wheat were found to be positively related to aphid densities (Harwood et al. 2004). Considering alternative prey, Harwood et al. (2004) found that Erigoninae spider aphid consumption was negatively influenced by Collembola densities but aphid predation rates by Linyphiinae were not affected by Collembola. The influence of hunting behavior by the two spider groups was explored by determining aphid consumption in web-based (sit-and wait) and non-web (active hunting) spiders. Harwood et al. (2004) suggested that higher densities of Collembola at Erigoninae web sites in contrast to non-web sites (Harwood et al. 2001) allowed the web-based Erigoninae to easily exploit Collembola while active hunters may be encountering aphids more often than web-based spiders. In the same study (Harwood et al. 2004), it was shown that Diptera abundance did not affect aphid consumption by either spider group. These findings with small, generally web-based, spiders contrast somewhat with our results using larger wolf spider that actively hunt prey.
In our study, the probability of detecting R. padi DNA in individual spiders differed significantly among farms, but was not positively related to R. padi density (Table 2). Instead, it was observed that R. padi consumption was affected by the presence of alternative prey; there was a positive association with the availability of alternative prey. In a previous study, based on gut-content analysis of spiders collected in fields with consistently low aphid densities, we found a negative association between R. padi consumption and Collembola availability (Kuusk and Ekbom 2010). As Pardosa spiders did not feed on Collembolans in relation to availability we suggested that the underlying mechanism was that the spiders changed their hunting behavior (from active to passive sit-and-wait) when mobile Collembolans became more numerous resulting in fewer encounters with sedentary aphids. To explain why approximately the same proportion of spiders tested positive for Collembola consumption at each sampling occasion irrespective of prey availability we suggested that the nutritional value of one or several species of the present Collembolans was of such importance that a fixed proportion of spiders regularly fed on them.
The positive relationship between R. padi consumption and alternative prey availability observed here contrasts with our previous findings. We suggest that the opposite relationships could be explained by the fact that in this study, densities of R. padi varied between sampling occasions and reached considerably higher levels than in the former, and because the spatial distribution of the prey was different. At high densities, aphids tend to leave the cereal plants and walk on the soil surface (Wiktelius and Ekbom 1985; Sopp et al. 1987). In our earlier study, when aphid densities were low in all fields, R. padi were exclusively observed on the plants and no aphids were caught on the ground-attached sticky-traps. In this study, R. padi were distributed both on plants and on the ground. Again, we suggest that an increased availability of mobile prey (Collembolans and Dipterans) made the spiders hunt more passively. However, while this change in foraging behavior caused decreased encounter rates with aphids on plants in the earlier study it had no negative effect on R. padi predation in the current as the prey items were available on the ground as well. Spiders may have consumed aphids more readily as they would be easier to catch than jumping springtails and flying Dipterans.
While spiders appeared to readily eat R. padi, fewer individuals than expected contained Collembola DNA with the exception of one sampling occasion Farm C June 30 (Fig. 1). In general, Collembola consumption was lower (~5–20 % positives) compared to R. padi consumption (~50–85 % positives) but on one sampling occasion (Farm C June 21) as many as 62 % of the spiders tested positive for Collembola remains (Fig. 1). On this date, Collembola availability was clearly highest which in turn explains the observed positive relationship between Collembola detection and Collembola availability (Table 2). Collembolans have a wide global distribution and usually occur in high numbers in soils and leaf litter. Thus, they form a potential source of non-pest alternative food for generalist predators and indeed there are a growing number of DNA-based gut-content studies that confirm that generalist predators feed on Collembola in the field (Agustí et al. 2003; Chapman et al. 2010; King et al. 2011). We have previously demonstrated that Pardosa feed on Collembolans in agricultural fields. In the former study though, predation rates were less variable; ~15–30 % tested positive at each sampling occasion compared to ~5–60 % here. To explain why Collembola consumption was more variable and why Collembola predation was disproportionally low at five out of six sampling occasions we suggest that the current ratios of potential prey and/or the nutritional needs of the spiders “favored” a foraging behavior that generally increased predation on aphids and potentially Dipterans. Kruse et al. (2008) demonstrated that the flexible body, chelicerae and legs of lycosid spiders make them highly adapted to catch flying prey, such as Dipterans, compared to carabid beetles, which have stiff bodies and inflexible legs and mandibles. Carabid beetles were capable of catching Drosophila melanogaster (Meigen) only at low temperatures (5–10 °C) when the flies were able to fly just a few centimeters or not at all. In contrast, P. prativaga was observed to readily catch D. melanogaster also at high temperatures. Apart from having a suitable morphology for catching flies, Dipterans could be an important source of proteins for Pardosa spiders. Dipterans contain relatively high amounts of this macronutrient due to their flight musculature (Marden 1989; McLachlan and Neems 1996) and experimental studies have shown that Pardosa spiders can forage selectively for protein-rich food if previous diets had been protein poor (Mayntz et al. 2005).
When studying field predation rates using PCR-based gut-content analysis prey detection rates are important for comparisons between predators. For the aphid amplicon, we have demonstrated that the median detection period in Pardosa spiders is about 4 h (Kuusk et al. 2008). There is no precise estimate for the Collembola amplicon, but we carried out a feeding experiment where spiders were tested for Collembola DNA immediately and 24 h after ingestion of one Isotoma spp. (Kuusk and Ekbom 2010). The analysis showed that Collembola DNA could be detected in 11 out of 12 spiders (92 %) that were frozen immediately after prey ingestion. After 24 h, it was not possible to detect the remains in any of the 12 fed spiders. The reason for testing the spiders after 24 h was that the sticky traps in the fields were exposed to prey for 24 h and we wanted to make sure that prey consumed before that period would not be detected. The feeding trials indicate that the detection times are similar and we argue that adjustments would not change the results in such a way that our conclusions would be different.
This study demonstrates that prey consumption by Pardosa spiders in agroecosystems cannot, in general, be assumed to simply mirror the ratio at which a particular food item is present in the field. When trying to obtain more knowledge about a predator’s foraging behavior other factors, such as the spatial distribution of the target prey and the abundance, composition, and nutritional content of potential alternative food items, need to be considered as well.
References
Agustí N, Shayler SP, Harwood JD, Vaughan IP, Sunderland KD, Symondson WOC (2003) Collembola as alternative prey sustaining spiders in arable ecosystems: prey detection within predators using molecular markers. Mol Ecol 12:3467–3475
Beckerman AP, Petchey OL, Warren PH (2006) Foraging biology predicts food web complexity. PNAS 103:13745–13749
Chapman EG, Romero SA, Harwood JD (2010) Maximizing collection and minimizing risk: does vacuum suction sampling increase the likelihood for misinterpretation of food web connections. Mol Ecol Resour 10:1023–1033
Chen Y, Giles KL, Payton ME, Greenstone MH (2000) Identifying key cereal aphid predators by molecular gut analysis. Mol Ecol 9:1887–1898
Chiverton PA (1986) Predator density manipulation and its effect on populations of Rhopalosiphum padi (Hom.: Aphididae) in spring barley. Ann Appl Biol 109:49–60
Edgar WD (1969) Prey and predators of the wolf spider Lycosa lugubris. J Zool 159:405–411
Foelix RF (1996) Biology of spiders, 2nd edn. Oxford University Press, New York
Gagnon AÈ, Doyon J, Heimpel GE, Brodeur J (2011) Prey DNA detection success following digestion by intraguild predators: influence of prey and predator species. Mol Ecol Resour 11:1022–1032
Greenstone MH, Szendrei Z, Payton ME, Rowley DL, Coudron TC, Weber DC (2010) Choosing natural enemies for conservation biological control: use of the prey detectability half-life to rank key predators of Colorado potato beetle. Entomol Exp Appl 136:97–107
Harwood JD, Sunderland KD, Symondson WOC (2001) Living where the food is: web location in relation to prey availability in winter wheat. J Appl Ecol 38:88–99
Harwood JD, Sunderland KD, Symondson WOC (2004) Prey selection by linyphiid spiders: molecular tracking of the effects of alternative prey on rates of aphid consumption in the field. Mol Ecol 13:3549–3560
Harwood JD, Bostrom MR, Hladilek EE, Wise DH, Obrycki JJ (2007) An order-specific monoclonal antibody to Diptera reveals the impact of alternative prey on spider feeding behavior in a complex food web. Biol Control 41:397–407
Harwood JD, Yoo HJS, Greenstone MH, Rowley DL, O′Neil RJ (2009) Differential impact of adults and nymphs of a generalist predator on an exotic invasive pest demonstrated by molecular gut-content analysis. Biol Invasions 11:895–903
Hatteland BA, Symondson WOC, King RA, Skage M, Schander C, Solhøy T (2011) Molecular analysis of predation by carabid beetles (Carabidae) on the invasive Iberian slug Arion lusitanicus. Bull Entomol Res 101:675–686
Hopkin SP (1997) Biology of the springtails (Insecta: Collembola). Oxford University Press, Oxford
Hosmer DW, Lemeshow S (2000) Applied logistic regression, 2nd edn. Wiley, New York
Juen A, Traugott M (2007) Revealing species-specific trophic links in soil food webs: molecular identification of scarab predators. Mol Ecol 16:1545–1557
King RA, Read DS, Traugott M, Symondson WOC (2008) Molecular analysis of predation: a review of best practice for DNA-based approaches. Mol Ecol 17:947–963
King RA, Vaughan JP, Bell JR, Bohan DA, Symondson WOC (2010) Prey choice by carabid beetles feeding on an earthworm community analysed using species- and lineage-specific PCR primers. Mol Ecol 19:1721–1732
King RA, Moreno-Ripoll R, Agustí N, Shayler SP, Bell JR, Bohan DA, Symondson WOC (2011) Multiplex reactions for the molecular detection of predation on pest and nonpest invertebrates in agroecosystems. Mol Ecol Resour 11:370–373
Kruse PD, Toft S, Sunderland KD (2008) Temperature and prey capture: opposite relationships in two predator taxa. Ecol Entomol 33:305–312
Kuusk A-K, Agustí N (2008) Group-specific primers for DNA-based detection of springtails (Hexapoda: Collembola) within predator gut contents. Mol Ecol Resour 8:678–681
Kuusk A-K, Ekbom B (2010) Lycosid spiders and alternative food: feeding behavior and implications for biological control. Biol Control 55:20–26
Kuusk A-K, Cassel-Lundhagen A, Kvarnheden A, Ekbom B (2008) Tracking aphid predation by lycosid spiders in spring-sown cereals using PCR-based gut-content analysis. Basic Appl Ecol 9:718–725
Leather SR, Walters KFA, Dixon AFG (1989) Factors determining the pest status of the bird cherry-oat aphid, Rhopalosiphum padi (L.) (Hemiptera: Aphididae), in Europe: a study and review. Bull Entomol Res 79:345–360
Marden JH (1989) Bodybuilding dragonflies: costs and benefits of maximizing flight muscle. Physiol Zool 62:505–521
Mayntz D, Raubenheimer D, Salomon M, Toft S, Simpson SJ (2005) Nutrient-specific foraging in invertebrate predators. Science 307:111–113
McLachlan AJ, Neems RM (1996) Is flight architecture determined by physical constraints or by natural selection? The case of the midge Chironomus plumosus. J Zool 240:301–308
Moser SE, Kajita Y, Harwood JD, Obrycki JJ (2011) Evidence for utilization of Diptera in the diet of field-collected coccinellid larvae from an antibody-based detection system. Biol Control 58:248–254
Öberg S, Ekbom B, Bommarco R (2007) Influence of habitat type and surrounding landscape on spider diversity in Swedish agroecosystems. Agric Ecosyst Environ 122:211–219
Öberg S, Cassel-Lundhagen A, Ekbom B (2011) Pollen beetles are consumed by ground- and foliage-dwelling spiders in winter oilseed rape. Entomol Exp Appl 138:256–262
Oelbermann K, Scheu S (2002) Effects of prey type and mixed diets on survival, growth and development of a generalist predator, Pardosa lugubris (Araneae: Lycosidae). Basic Appl Ecol 3:285–291
Östman Ö, Ekbom B, Bengtsson J (2001) Landscape heterogeneity and farming practice influence biological control. Basic Appl Ecol 2:365–371
Östman Ö, Ekbom B, Bengtsson J (2003) Yield increase attributable to aphid predation by ground-living polyphagous natural enemies in spring barley in Sweden. Ecol Econ 45:149–158
Samu F, Szinetár C (2002) On the nature of agrobiont spiders. J Arachnol 30:389–402
Samu F, Sziranya A, Kiss B (2003) Foraging in agricultural fields: local sit-and-move strategy scales up to risk-averse habitat use in a wolf spider. Anim Behav 66:939–947
Schmidt MH, Roschewitz I, Thies C, Tscharntke T (2005) Differential effects of landscape and management on diversity and density of ground-dwelling farmland spiders. J Appl Ecol 42:281–287
Schmitz OJ (2007) Predator diversity and trophic interactions. Ecology 88:2415–2426
Sheppard SK, Harwood JD (2005) Advances in molecular ecology: tracking trophic links through predator–prey food-webs. Funct Ecol 19:751–762
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:652–701
Sopp PI, Sunderland KD, Coombes DS (1987) Observations on the number of cereal aphids on the soil in relation to aphid density in winter wheat. Ann Appl Biol 111:53–57
Symondson WOC, Sunderland KD, Greenstone MH (2002) Can generalist predators be effective biological control agents? Ann Rev Entomol 47:561–594
Tahir HM, Butt A (2009) Predatory potential of three hunting spiders inhabiting the rice ecosystems. J Pest Sci 82:217–225
Toft S (1995) Value of the aphid Rhopalosiphum padi as food for cereal spiders. J Appl Ecol 32:552–560
Toft S (1997) Acquired food aversion of a wolf spider to three cereal aphids: intra- and interspecific effects. Entomophaga 42:63–69
Wiktelius S (1987) Distribution of Rhopalosiphum padi (Homoptera, Aphididae) on spring barley plants. Ann Appl Biol 110:1–7
Wiktelius S, Ekbom BS (1985) Aphids in spring sown cereals in central Sweden. Abundance and distribution 1980–1983. Z Ang Entomol 100:8–16
Acknowledgments
This project was financed by the research council FORMAS. We thank the farmers who allowed us to work in their fields; Martina Sjöström and Hernán Guzmán who assisted in collecting spiders; Carol Högfeldt and Gerard Malsher for help in the lab; and Michael Andersson for statistical advice. We also thank the anonymous referees for providing useful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Traugott.
Rights and permissions
About this article
Cite this article
Kuusk, AK., Ekbom, B. Feeding habits of lycosid spiders in field habitats. J Pest Sci 85, 253–260 (2012). https://doi.org/10.1007/s10340-012-0431-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-012-0431-4