Abstract
In 2002, a powdery mildew with catenate conidia lacking fibrosin bodies was found on cucumber in a greenhouse in Kanagawa Prefecture, Japan. Morphological observation revealed that the fungus belongs to Oidium subgenus Reticuloidium, anamorph of the genus Golovinomyces. Molecular phylogenetic analyses of the nucleotide sequences of the rDNA ITS regions and D1/D2 domains of the 28S rDNA indicated that the fungus belongs to the clade of G. orontii with other Golovinomyces fungi from a wide range of host plants, suggesting that the fungus was newly transported from abroad. Because there has been no prior report of cucumber powdery mildew caused by Reticuloidium, further research on the physiology, epidemiology, control and resistant cucumber varieties is required.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Phytopathological Society of Japan (2000) recorded four powdery mildew fungi on Cucumis sativus in Japan: Erysiphe polygoni, Oidiopsis sicula (anamorph of Leveillula taurica; Saito and Kurata 1975), Oidium sp. of polygoni-type (Oidium subgenus Pseudoidium; Sato et al. 1996), and Sphaerotheca cucurbitae (=Podosphaera xanthii; anamorph: Oidium subgenus Fibroidium). Because the anamorph of E. polygoni belongs to Oidium subgenus Pseudoidium, the powdery mildews of C. sativus may be reduced to three. Because the occurrence of Oidiopsis sicula and Oidium subgenus Pseudoidium has been reported only once in Japan, the major causal agent of the cucumber powdery mildew seems to be S. cucurbitae. Based on morphological observations of the conidial stage and on molecular phylogenetic analyses, the genus Sphaerotheca was merged into the genus Podosphaera (Braun and Takamatsu 2000), and the name of the cucumber mildew was revised to Podosphaera xanthii (Braun et al. 2001). We therefore named the Podosphaera species on cucumber in the present study as Podosphaera xanthii and its anamorph as Fibroidium.
In 2002, three powdery mildews with conidial stages of different morphologies were found on cucumber plants in a greenhouse in Hiratsuka-shi, Kanagawa Prefecture, Japan. Two of the fungi were identified as Oidium subgenus Fibroidium and Oidium subgenus Pseudoidium. The third mildew with catenate conidia and lacking fibrosin bodies belonged to Oidium subgenus Reticuloidium, anamorph of the genus Golovinomyces. Golovinomyces orontii is a common powdery mildew of cucurbits worldwide, including Europe, North America, South America, Africa, and West and Central Asia (Amano 1986). In East Asia, although E. cucurbitacearum (=G. orontii) has been reported to occur in China, there is no record of this fungus in Korea and Japan (Braun 1987; Shin 2000; Phytopathological Society of Japan 2000). Therefore, we characterized the fungus through morphological observations and molecular phylogenetic analyses.
Materials and methods
Fungal isolation
Leaf disks with a diameter of 9 mm were cut from cotyledons of C. sativus (cv. Tokiwa Hikari-3P) with a cork borer. One hundred leaf disks were put on a wet filter paper in a Petri dish (150 mm in diameter) and exposed for 3 h at two locations in a greenhouse with diseased cucumber plants. Then, the leaf disks were moved to small Petri dishes (60 mm in diameter; a single leaf/dish) and incubated for 6 days at 20°C under 2,000–2,500 lux with a photoperiod of 12/12 h (day/night) in an incubation room. A single spore was picked up with an endodontic file from a colony that developed on the leaf disk and placed on a healthy cucumber cotyledon, then incubated under the same conditions. Powdery mildew of Lamium amplexicaule in the same greenhouse, was also isolated by the same procedure. Cucumber seedlings (cultivar Tokiwa Hikari-3P) at the 2–3 leaf stage were sprayed with conidia of the isolates every 2 weeks and kept in a plastic box (20 × 20 × 30 cm) until the pathogenicity test.
Morphological observations
Hyphae, conidiophores and conidia were stripped off the surface of a detached leaf with clear adhesive tape, mounted on a microscope glass slide with the fungal mycelium uppermost and examined in water. To observe conidial germ tubes, we inoculated the inner cell layer of onion scales using the method of Hirata (1942, 1955).
Pathogenicity test
An isolate of Fibroidium B-7SF and two isolates of Reticuloidium No. 2–7 (from cucumber) and B-7H2 (from Lamium amplexicaule) were used to test pathogenicity. Potted cucumber plants (cvs. Sharp 1, Haru-no Megumi and Tokiwa Hikari-3P) were grown in Kureha horticultural compost (Kureha Chemical, Tokyo, Japan) at 25°C under 3,000–5,000 lux with a photoperiod of 12/12 h (day/night) in a phytotron for 2–3 weeks. These plants were then sprayed with a conidial suspension [ca. 105 conidia/ml, amended with 1/5,000 (v/v) Tween 20] of an isolate. The inoculated plants were put into a plastic box and incubated for 21 days at 20°C under 2,000–2,500 lux at the photoperiod of 12/12 h (day/night). The percentage of infected area of each of the powdery-mildewed cucumber leaves was determined by the following scale: 0 = no colony, 0.5 = percentage of area infection ≤10%, 1 = >10–20%, 2 = >20–40%, 3 = >40–60%, 4 = >60–80%, 5 = >80%. Disease severity was calculated as [(5A + 4B + 3C + 2D + E + 0.5F)/5G] × 100, where A, B, C, D, E and F are the number of leaves corresponding to the scale at 5, 4, 3, 2, 1 and 0.5, respectively, and G is the total number of leaves assessed.
Molecular phylogenetic analyses
Whole-cell DNA was extracted from mycelia by the chelex method (Walsh et al. 1991) as described in Hirata and Takamatsu (1996). The rDNA ITS region and the 5′ end of the 28S rDNA, including the D1 and D2 regions, were separately amplified two times by a polymerase chain reaction using nested primer combinations, and then sequenced directly as described in Matsuda and Takamatsu (2003).
The sequences were initially aligned using the Clustal_X program (Thompson et al. 1997) and further analyzed to improve the alignment with a word processing program with colour-coded nucleotides and deposited in TreeBASE (http://www.treebase.org/) with accession number S2265. Phylogenetic trees were obtained from the data by the maximum-parsimony method using the heuristic search option in the program PAUP* 4.0b8 (Swofford 2001). This search was repeated 100 times with different random starting points, using the stepwise addition option to increase the likelihood of finding the most parsimonious tree. Transversions and transitions were treated as equal weight. All sites were treated as unordered, with gaps treated as missing data. The branch-swapping algorithm was the tree bisection and reconstruction method, the MulTrees option was in effect, and zero-length branches were collapsed. The strength of the internal branches from the resulting trees was tested by bootstrap analysis (Felsenstein 1985) using 1,000 replications.
Results
Morphological observation
To investigate the powdery mildew species occurring in a greenhouse of cucumber of Hiratsuka-shi, Kanagawa Prefecture, we examined all 106 powdery mildew colonies that appeared on 52 naturally infected leaves of 80 cucumber plants (cv. Sharp-1) in the greenhouse in June 2002. As a result, 83, 22 and 1 colonies were identified as Oidium subgenera Reticuloidium, Fibroidium and Pseudoidium, respectively, based on Braun et al. (2002). The morphology of Reticuloidium was as follows: white mycelium on leaves, amphigenous, powdery-like. Leaf surface with colony slightly swelled out (Fig. 1a). Conidiophores erect, foot-cells slightly curved at the base, conidia produced in chains following 2–4 mother cells, semicylindrical or doliform, germ tubes arising from the middle portion of the conidia, tips unlobed, slightly enlarged with nipple-shaped appressoria (cichoracearum-type) (Table 1; Fig. 1c–e). The morphology of single-spore isolate No. 2–7 was consistent with these characteristics.
Powdery mildews also occurred on Lamium amplexicaule (Fig. 1b) and Oxalis carniculata, common weeds both inside and outside of the greenhouse where cucumber powdery mildew was found. Powdery mildew on Oxalis carniculata belonged to subgenus Pseudoidium based on the characteristics of having noncatenate conidia and polygoni-type conidial germ tubes and is regarded as an anamorph of Erysiphe russellii (Nomura 1997). The fungus on Lamium amplexicaule belonged to subgenus Reticuloidium, from which a single-spore isolate, B-7H2, was established (Table 1).
Pathogenicity test
Reticuloidium as well as Fibroidium are pathogenic on the three cucumber varieties used in this study. However, the disease severities with Reticuloidium isolates No. 2–7 (disease severity = 16–45%) and B-7H2 (22–42%) were lower than that of Fibroidium isolate B-7SF (64–83%) (Table 2). The latent period of Reticuloidium was about 7 days longer than that of Fibroidium . In this study, we estimated disease severity 21 days after inoculation. The different disease severities between Fibroidium and Reticuloidium may be derived from the different latent period of these fungi. Reticuloidium isolate No. 2–7 from cucumber was pathogenic on Lamium amplexicaule.
ITS phylogeny
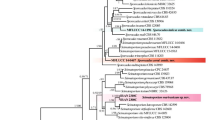
The rDNA ITS region and the D1/D2 domains of the 28S rDNA were sequenced for each isolate from C. sativus (cv. No. 1–16) and Lamium amplexicaule (cv. B-7H2). The sequences were deposited in the DDBJ DNA database with accession numbers AB427187 and AB427188, respectively. These two sequences were identical in both ITS and 28S rDNA regions. The two ITS sequences obtained in this study were aligned with 70 ITS sequences retrieved from the DDBJ database. The ITS data set consists of 72 taxa, of which 70 are members of the genus Golovinomyces (including Oidium subgenus Reticuloidium). Of the 70 Golovinomyces sequences, five sequences (shown by solid circle in Fig. 2) are from cucurbit powdery mildew. The ITS sequences of the isolates from C. sativus and Lamium amplexicaule were identical to the sequences of G. orontii from Arabidopsis thaliana (AF031282) and Veronica arvensis (AB077652) and differed by one base from the sequences of G. orontii from Cucurbita pepo (AF229017 and AB077670), Cucurbita sp. (AB077696), Nicotiana tabacum (AB022413); G. cichoracearum from Sonchus sp. (AB077669), Cichorium intybus (AB077695, AB077666 and AF011294), Lactuca serriola (AB077688), and Mycelis muralis (AB077661); and Oidium sp. on Dahlia pinnata (AB077677).
Phylogenetic analysis of the nucleotide sequences of the ITS region including the 5.8S rDNA for 70 sequences from the genus Golovinomycetes (including Oidium subgenus Reticuoidium) and two outgroup sequences. The tree is a phylogram of the maximum likelihood tree among the 640 most parsimonious trees with 303 steps, which was obtained by a heuristic search employing the random stepwise addition option in the program PAUP*. Gaps were treated as missing data. Horizontal branch lengths are proportional to the number of nucleotide substitutions that were inferred to have occurred along a particular branch of the tree. Percentage bootstrap support (1,000 replications; ≥70%) are shown on the branches. Solid circle before a taxon label indicates a sequence from a cucurbit powdery mildew
The data set consisted of 534 characters, from which 39 sites were removed to circumvent ambiguous alignments. Of the 495 remaining characters, 138 sites were variable, and 116 sites were phylogenetically informative for parsimony analyses. Two sequences from Arthrocladiella mougeotii were used as the outgroup based on Mori et al. (2000). The parsimony analysis using PAUP* generated 640 equally parsimonious trees of 303 steps (consistency index [CI] = 0.6469, retention index [RI] = 0.8981, rescaled consistency index [RC] = 0.5809). One of the 640 trees with the highest log-likelihood value is shown in Fig. 2, which shows nine clades differentiated within the Golovinomyces taxa investigated in this study. Each clade was strongly supported by bootstrap values of 94% or more. Of the nine clades, four clades, I, V, VI and VII, comprise isolates from each of the host tribes of the Asteraceae: clade I from tribe Cardueae, clade V from Anthemideae, clade VI from Astereae, and clade VII from Heliantheae. Clade II comprises isolates from Lamiaceae and Ranunculaceae. Clade III comprises a single isolate from Lapsana (Asteraceae), clade IV comprises isolates from Lycopersicon and Solanum (Solanaceae), and clade VIII comprises isolates from Phlox (Polemoniaceae) and Physalis (Solanaceae). Clade IX is large and comprises isolates from the tribe Lactuceae of the Asteraceae, and many other plant families such as Plantaginaceae, Solanaceae, Brassicaceae and Cucurbitaceae. Sequences from the isolates No. 1–16 (from C. sativus) and B-7H2 (from Lamium amplexicaule) belonged to the clade IX. All the nine clades were commonly appeared in all 640 parsimonious trees, although the branching order of the clades differed among the trees.
28S phylogeny
The two 28S rDNA sequences obtained in this study were aligned with 49 28S rDNA sequences retrieved from the DDBJ database. The 28S rDNA data set consists of 51 sequences, of which 50 are from members of the genus Golovinomyces. These 50 sequences include 11 Golovinomyces species from 44 different plant species that represent 13 plant families, of which three sequences shown by solid circle in Fig. 3 are from cucurbit powdery mildews. The two 28S rDNA sequences determined in this study differed one base from G. cichoracearum on Lactuca serriola (AB077687) and Picris hietacioides (AB077686), and two bases from G. cichoracearum on Sonchus sp. (AB077668), Cichorium intybus (AB077667), Mycelis muralis (AB077660) and Lactuca serriola (AB077651), and G. orontii on Veronica arvensis (AB077651). There was no sequence identical to these two 28S rDNA sequences.
Phylogenetic analysis of the divergent domains D1 and D2 sequences of the 28S rDNA for 50 sequences from the genus Golovinomycetes (including Oidium subgenus Reticuloidium) and one outgroup sequence. The tree is one of 48,109 most parsimonious trees with 121 steps and was obtained by a heuristic search using PAUP*. Gaps were treated as missing data. Horizontal branch lengths are proportional to the number of nucleotide substitutions that were inferred to have occurred along a particular branch of the tree. Percentage bootstrap support (1,000 replications; ≥70%) is shown on the branches. Solid circle before a taxon label indicates a sequence from a cucurbit powdery mildew
The data set consists of 718 characters, all of which were used for tree construction because all their sites were aligned unambiguously. Of the 718 characters, 65 sites were variable, and 45 were phylogenetically informative for parsimony analyses. A sequence from Arthrocladiella mougeotii was used as an outgroup based on Mori et al. (2000). The parsimony analysis using PAUP* generated 48,109 equally parsimonious trees of 121 steps (CI = 0.6694, RI = 0.8964, RC = 0.6001). Because the tree topologies were almost identical among the 48,109 trees except for branching order of terminal branches, one of the trees is shown in Fig. 3. All trees support the respective clades recognized in the ITS analysis, although clades III and IV were not included in the 28S tree due to the lack of DNA sequence data. The two sequences from isolates No. 1–16 (from C. sativus) and B-7H2 (from Lamium amplexicaule) belonged to the large terminal clade IX.
Discussion
Four powdery mildew species, i.e., Erysiphe cichoracearum (=E. cucurbitacearum, E. polyphaga; present name: G. orontii; anamorph: Oidium subgenus Reticuloidium), E. communis (anamorph: Oidium subgenus Pseudoidium), Sphaerotheca fuliginea (present name: Podosphaera xanthii; anamorph: Oidium subgenus Fibroidium) and Leveillula taurica (anamorph: Oidiopsis siculae) are listed as causal agents of cucumber powdery mildew in the world (Amano 1986). Three powdery mildews, i.e., Oidium subgenus Pseudoidium, Podosphaera xanthii (=S. fuliginea, S. fusca, S. cucurbitae) and Leveillula taurica, have been reported to occur in Japan (Phytopathological Society of Japan 2000; Saito and Kurata 1975; Sato et al. 1996). Of these, Podosphaera xanthii is the main causal agent and the other two fungi are rare. The fungus from Hiratsuka-shi morphologically resembled Podosphaera xanthii in producing catenate conidia, but differed from Oidium subgenus Pseudoidium and Leveillula taurica, which have noncatenate conidia. However, the present fungus was also distinguished from Podosphaera xanthii in not having distinct fibrosin bodies in its conidia. In addition, the conidial germ tube of Podosphaera xanthii is the fuliginea-type, whereas that of the present fungus is the cichoracearum-type. These characteristics reveal that the present fungus differs from Podosphaera xanthii and belongs to Oidium subgenus Reticuloidium. This is the first record of cucumber powdery mildew caused by Reticuloidium in Japan. Because the teleomorph of Reticuloidium belongs to the genus Golovinomyces (Braun et al. 2002), the present fungus may be an anamorphic stage of G. orontii, a common powdery mildew of cucumber in the world except East Asia (Sitterly 1978; Braun 1987). Results of the molecular analysis showing that the fungus belongs to the clade of G. orontii support the assumption.
Reticuloidium mildew on cucumber was classified as E. cichoracearum in the monograph of Salmon (1900). E. cichoracearum sensu Salmon was reported from a wide range of plant families including Asteraceae, but considered as a complex of numerous races with narrow host ranges. Hammarlund (1945) inoculated 99 plant species from a wide range of families with an ‘E. cichoracearum’ isolate from cucumber and reported that the fungus infected 62 plant species of 11 families such as Asteraceae, Crassulaceae, Cucurbitaceae, and Solanaceae. Thus, he concluded that this fungus has a wide host range, although it resembled E. cichoracearum in morphology, and called the fungus ‘E. polyphaga’. Blumer (1952, 1967) performed independent inoculation tests and confirmed the presence of such a fungus with a wide host range. Braun (1980) found that the foot cells of the conidiophore of ‘E. polyphaga’ often curved at the base and proposed the name ‘E. orontii’ (the oldest available name for this fungus). He confined the name ‘E. cichoracearum’ to fungi on Asteraceae. The genus Erysiphe was then split into three genera based on the morphology of the anamorph and on molecular analyses, and E. orontii was revised as G. orontii (Braun 1999; Heluta 1988).
We used five cucurbitaceous mildew sequences in the present phylogenetic analysis. There is no sequence identical to the ITS sequence of the present Japanese isolates. AF229017 sequenced in France differs by only one base from the Japanese isolate at the 125th site from 5′-end of the ITS1 region. Recently, a new occurrence of Reticuloidium has been reported on several cultivated plants such as Helianthus tuberosus, Viola tricolor and Zinnia elegans (Hoshi et al. 2007; Tanda and Suga 2002). It is likely that these Reticuloidium mildews were newly imported from abroad.
We found Reticuloidium on Lamium amplexicaule in the same greenhouse in which cucumber Reticuloidium was found. Both the ITS and the 28S rDNA sequences of this isolate are identical to those of the cucumber mildew, and the isolate is pathogenic on cucumber. This result suggests that Lamium amplexicaule provided the primary inoculum for the cucumber mildew in this greenhouse. Neoerysiphe galeopsidis has been the only powdery mildew species to infect Lamium species (Amano 1986; Braun 1987). Blumer (1952) reported that Reticuloidium infects Lamium galeobdolon, and Garibaldi et al. (2007) reported the occurrence of G. orontii on Lamium galeobdolon. Uchida et al. (2002) found Oidium subgenus Fibroidium on Lamium amplexicaule near the cucumber greenhouses in Hiratsuka-shi. This Fibroidium was pathogenic to both cucumber and Lamium amplexicaule, suggesting that common weeds like Lamium amplexicaule and Lamium purpureum serve as hosts to produce primary inoculum of Fibroidium as well as Reticuloidium to cause cucumber mildew.
In Europe and other regions of the world, G. orontii is an important causal agent of cucurbit mildew with Podosphaera xanthii (Bardin et al. 1999; Jahn et al. 2002; Mohamed et al. 1995; Sitterly 1978; Sz Nagy 1970, 1972, 1976; Vakalounakis and Klironomou 1995; Vakalounakis et al. 1994). Based on Sz Nagy (1976), ‘E. cichoracearum’ (=Reticuloidium) can germinate in wider temperature ranges than ‘Sphaerotheca fuliginea’ (=Podosphaera xanthii; Fibroidium), and conidia of the former species are able to germinate at lower humidity. Hoshi et al. (2007) reported that Reticuloidium occurs earlier than Fibroidium in Japan. Thus, the two mildew fungi differ in some physiological and ecological characteristics. Although evidence on cucumber mildew caused by Fibroidium has been accumulating for many years in Japan (Abiko 1978, 1982a, 1982b; Endo 1989; Hosoya et al. 1999, 2000; Kuzuya et al. 2003, 2006), there is no report of cucumber mildew caused by Reticuloidium. Further research on the physiology, epidemiology, chemical control of this fungus and resistant varieties of cucumber is needed.
References
Abiko K (1978) Studies on the specialization of parasitism of Sphaerotheca fuliginea (Schlecht.) Pollacci I. Powdery mildew fungi parasitic on cucurbits, eggplant, edible burdock and Japanese butterbur. Ann Phytopathol Soc Jpn 44:612–618
Abiko K (1982a) Studies on the specialization of parasitism of Sphaerotheca fuliginea (Schlecht.) Pollacci II. Powdery mildew fungi on flowering plants. Bull Veg Ornam Crops Res Stn Ser A 10:57–62
Abiko K (1982b) Studies on the specialization of parasitism of Sphaerotheca fuliginea (Schlecht.) Pollacci II. Powdery mildew fungi parasitic on weeds. Bull Veg Ornam Crops Res Stn Ser A 10:63–67
Amano K (1986) Host range and geographical distribution of the powdery mildew fungi. Japan Scientific Societies Press, Tokyo
Bardin M, Carlier J, Nicot PC (1999) Genetic differentiation in the French population of Erysiphe cichoracearum, a causal agent of powdery mildew of cucurbits. Plant Pathol 48:531–540
Blumer S (1952) Beiträge zur Spezialisation der Erysipheen (in German). Ber Schweiz Bot Ges 62:384–401
Blumer S (1967) Echte Mehltaupilze (Erysiphaceae) (in German). Gustav Fischer, Jena
Braun U (1980) Morphological studies in the genus Oidium. Flora 170:77–90
Braun U (1987) A monograph of the Erysiphales (powdery mildews). Beih Nova Hedwig 89:1–700
Braun U (1999) Some critical notes on the classification and generic concept of the Erysiphaceae. Schlechtendalia 3:49–55
Braun U, Takamatsu S (2000) Phylogeny of Erysiphe, Microsphaera, Uncinula (Erysipheae) and Cystotheca, Podosphaera, Sphaerotheca (Cystotheceae) inferred from rDNA ITS sequences—some taxonomic consequences. Schlechtendalia 4:1–33
Braun U, Shishkoff N, Takamatsu S (2001) Phylogeny of Podosphaera sect. Sphaerotheca subsect. Magnicellulatae (Sphaerotheca fuliginea auct. s. lat.) inferred from rDNA ITS sequences—a taxonomic interpretation. Schlechtendalia 7:45–52
Braun U, Cook RTA, Inman AJ, Shin HD (2002) The taxonomy of the powdery mildew fungi. In: Bélanger R, Bushnell WR, Dik AJ, Carver TLW (eds) The powdery mildews: a comprehensive treatise. APS Press, St. Paul, pp 13–55
Endo T (1989) Studies on the life-cycle of cucurbit powdery mildew fungus Sphaerotheca fuliginea (Schlecht.) Poll (in Japanese with English summary). Spec Bull Fukushima Pref Agr Exp Stn 5:1–106
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Garibaldi A, Bertetti D, Minerdi D, Gullino ML (2007) First report of powdery mildew caused by Golovinomyces orontii (Erysiphe orontii) on Lamium galeobdolon in Italy. Plant Dis 91:635
Hammarlund C (1945) Beiträge zur Revision einiger imperfekter Mehltau-Arten. Erysiphe polyphaga nov. sp. (in German). Bot Not 1945:101–108
Heluta VP (1988) Phylogenetic connections among genera of powdery mildew fungi and some questions of systematics of Erysiphales. Biol J Arm 41:351–358
Hirata K (1942) On the shape of the germ tubes of Erysiphaceae (in Japanese). Bull Chiba Coll Hort 5:34–49
Hirata K (1955) On the shape of the germ tubes of Erysiphaceae II (in Japanese). Bull Fac Agric Niigata Univ 7:24–36
Hirata T, Takamatsu S (1996) Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37:283–288
Hoshi H, Sato Y, Horie H (2007) Occurrence of zinnia and Jerusalem artichoke powdery mildew by Oidium subgenus Reticuloidium, and host range of the subgenus of powdery mildew on several plant species occurred at Tokyo (abstract in Japanese). Ann Phytopathol Soc Jpn 73:182
Hosoya K, Narisawa K, Pitrat M, Ezura H (1999) Race identification in powdery mildew (Sphaerotheca fuliginea)on melon (Cucumis melo L.) in Japan. Plant Breeding 118:259–262
Hosoya K, Kuzuya M, Murakami T, Kato K, Narisawa K, Ezura H (2000) Impact of the resistant melon cultivars on Sphaerotheca fuliginea. Plant Breeding 119:286–288
Jahn M, Munger HM, McCreight JD (2002) Breeding cucurbit crops for powdery mildew resistance. In: Bélanger R, Bushnell WR, Dik AJ, Carver TLW (eds) The powdery mildews: a comprehensive treatise. APS Press, St Paul, pp 239–248
Kuzuya M, Hosoya K, Yashiro K, Tomita K, Ezura H (2003) Powdery mildew (Sphaerotheca fuliginea) resistance in melon is selectable at the haploid level. J Exp Bot 54:1069–1074
Kuzuya M, Yashiro K, Tomita K, Ezura H (2006) Powdery mildew (Podosphaera xanthii) resistance in melon is categorized into two types based on inhibition of the infection processes. J Exp Bot 57:2093–2100
Matsuda S, Takamatsu S (2003) Evolution of host–parasite relationship of Golovinomyces (Ascomycete: Erysiphales) inferred from nuclear rDNA sequences. Mol Phylogenet Evol 27:314–327
Mohamed YF, Bardin M, Nicot PC, Pitrat M (1995) Causal agents of powdery mildew of cucurbits in Sudan. Plant Dis 79:634–636
Mori Y, Sato Y, Takamatsu S (2000) Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia 92:74–93
Nomura Y (1997) Taxonomical study of Erysiphaceae of Japan (in Japanese with English summary). Yokendo, Tokyo
Phytopathological Society of Japan (ed) (2000) Common names of plant diseases in Japan (in Japanese). Japan Plant Protection Association, Tokyo
Saito M, Kurata M (1975) Occurrence of Leveillula taurica on cucumber, okra and eggplant (abstract in Japanese). Ann Phytophathol Soc Jpn 41:269
Salmon E (1900) A monograph of the Erysiphaceae. Mem Torrey Bot Club 9:1–292
Sato Y, Nakamura T, Takamatsu S, Morikawa T, Chikuo Y (1996) Powdery mildew fungi newly found on Datura stramonium and Cucumis sativus in Japan (abstract in Japanese). Ann Phytophathol Soc Jpn 62:630
Shin HD (2000) The Erysiphaceae of Korea. National Institute of Agricultural Science & Technology, Suwon
Sitterly WR (1978) Powdery mildews of cucurbits. In: Spencer DM (ed) The powdery mildews. APS Press, St. Paul, pp 359–379
Swofford DL (2001) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b8. Sinauer, Sunderland
Sz Nagy G (1970) Die Identifizierung des Mehltaus der Kürbisgewächse auf Grund der Konidienmerkmale (in German with English summary). Acta Phytopathol Hung 5:231–248
Sz Nagy G (1972) Studies on powdery mildews of cucurbits. I. Host range and maintenance of Sphaerotheca fuliginea and Erysiphe sp. under laboratory and glasshouse conditions. Acta Phytopathol Hung 7:415–420
Sz Nagy G (1976) Studies on powdery mildews of cucurbits II. Life cycle and epidemiology of Erysiphe cichoracearum and Sphaerotheca fuliginea. Acta Phytopathol Hung 7:415–420
Tanda S, Suga R (2002) Powdery mildews occurred on horseradish and five economic plants, and their causal fungi (in Japanese with English summary). J Agri Sci Tokyo Univ of Agric 47:141–152
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Uchida K, So K, Niinomi S, Takamatsu S (2002) Morphological characteristics and parasitism of powdery mildew on Lamium amplexicaule (abstract in Japanese). Ann Phytophathol Soc Jpn 68:189
Vakalounakis DJ, Klironomou E (1995) Race and mating type identification of powdery mildew on cucurbits in Greece. Plant Pathol 44:1033–1038
Vakalounakis DJ, Klironomou E, Papadakis A (1994) Species spectrum, host range and distribution of powdery mildews on Cucurbitaceae in Crete. Plant Pathol 43:813–818
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506–513
Acknowledgments
We are grateful to Dr. Hideo Ishii (National Institute for Agro-Environmental Sciences, Japan) for his helpful advice during this study and for critically reading the manuscript. This work was partially supported by a Grant-in-Aid for Scientific Research (C) (No. 18580046) from the Ministry of Education, Science, Sports and Culture of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Uchida, K., Takamatsu, S., Matsuda, S. et al. Morphological and molecular characterization of Oidium subgenus Reticuloidium (powdery mildew) newly occurred on cucumber in Japan. J Gen Plant Pathol 75, 92–100 (2009). https://doi.org/10.1007/s10327-009-0146-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-009-0146-4