Abstract
Diesel fuel is one of the most important sources of hydrocarbon contamination worldwide. Its composition consists of a complex mixture of n-alkanes, branched alkanes and aromatic compounds. Hydrocarbon degradation in Pseudomonas species has been mostly studied under aerobic conditions; however, a dynamic spectrum of oxygen availability can be found in the environment. Pseudomonas extremaustralis, an Antarctic bacterium isolated from a pristine environment, is able to degrade diesel fuel and presents a wide microaerophilic metabolism. In this work RNA-deep sequence experiments were analyzed comparing the expression profile in aerobic and microaerophilic cultures. Interestingly, genes involved in alkane degradation, including alkB, were over-expressed in micro-aerobiosis in absence of hydrocarbon compounds. In minimal media supplemented with diesel fuel, n-alkanes degradation (C13–C19) after 7 days was observed under low oxygen conditions but not in aerobiosis. In-silico analysis of the alkB promoter zone showed a putative binding sequence for the anaerobic global regulator, Anr. Our results indicate that some diesel fuel components can be utilized as sole carbon source under microaerophilic conditions for cell maintenance or slow growth in a Pseudomonas species and this metabolism could represent an adaptive advantage in polluted environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial adaptability to different environmental conditions requires different strategies, including the response of individual genes or operons and complex regulatory networks that coordinate the control of several genes [25]. Oxygen availability is a key factor in bacterial physiology; in the environment, the uneven distribution of water flow, nutrients, and microbial populations creates a dynamic spectrum of aerobic, microaerophilic, and anaerobic conditions [9]. The heterogeneity of oxygen distribution requires physiological responses that could include the capability to use different electron acceptors as well as the fermentation of carbon compounds [32].
Hydrocarbon contamination has become a tough problem worldwide. One of the most widely distributed sources of this kind of contamination is diesel fuel, a complex mixture of n-alkanes, branched alkanes and aromatic hydrocarbons. Due to the lack of functional groups and low water solubility, aliphatic hydrocarbons exhibit both low chemical reactivity and bioavailability for microorganism [7]. Pseudomonas species are capable of using n-alkanes as carbon source by activating the hydrocarbon as a key first step [5]. This reaction is mostly driven by the enzyme 1-alkane monooxygenase encoded by alkB [11]. It has been described that degradation of n-alkanes is initiated by the oxidation of a terminal methyl group to render a primary alcohol, which gets further oxidized to an aldehyde, and finally converted into a fatty acid [30], which is finally processed by β-oxidation to generate acetyl-CoA [42]. This process has been described to occur under aerobic conditions especially in Pseudomonas species [31].
Pseudomonas extremaustralis is a bacterium isolated from Antarctica that shows high stress resistance in association with the production of high amounts of polyhydroxyalkanoates [23]. This strain is capable of growing and developing biofilms under low temperatures and survives freezing [3]. Additionally, the microaerophilic metabolism of P. extremaustralis has been studied and the role of the anaerobic global regulator, Anr, has been analyzed in polyhydroxybutyrate metabolism, redox state, oxidative stress resistance under low oxygen conditions and biofilm development [35, 37, 38]. It was also described that P. extremaustralis was able to grow using diesel fuel as sole carbon source only when cultured in biofilm [36].
RNA-deep sequencing is a powerful tool for analyzing gene expression and has been used to study the transcriptome profile under different conditions, for example, in P. aeruginosa PAO-1 under stress conditions, and P. putida KT2440 and P. extremaustralis under low temperature conditions [14, 15, 39].
In this work the alkane degradation pathway of P. extremaustralis at low oxygen tensions using both high throughput experiments and physiological assays in planktonic cultures was explored. The results demonstrate that 1-alkane monooxygenase is over-expressed under low oxygen conditions regardless of the carbon source, in line with the degradation of alkanes that occurs mainly under low oxygen conditions. This alkane degradation metabolism allows supporting bacterial survival in a newly contaminated environment where these slow growing microorganisms could be the first step in microbial succession. The obtained results provide new information that could be important to improve bioremediation strategies by the stimulation of these microorganisms when oxygen is a limiting factor as found after a recent oil spill.
Materials and methods
Strains and cultured conditions
Pseudomonas extremaustralis—a bacterium isolated from a temporary pond in Antarctica [23] was used through the experiments. Experiments under microaerophilic condition were carried out in hermetically sealed bottles using a 1:2 medium-to-flask volume ratio without shaking, while aerobic condition experiments were performed in shaken Erlenmeyers using 1:10 medium-to-flask volume ratio at 200 RPM. For RNA-seq experiments cultures were grown in LB medium supplemented with KNO3 (0.08%) at 30 °C. In hydrocarbon degradation experiments pre-inocula were grown overnight in LB medium supplemented with 0.25% sodium octanoate. These cultures were used to inoculate sealed bottles in E2 minimal medium [19] supplemented with 1% diesel fuel (YPF—Yacimientos Petroliferos Fiscales-Argentina) at an initial OD600nm of 0.05. When necessary, 0.05% glucose and/or 0.08% KNO3 were added. Cultures were incubated at 30 °C under microaerophilic conditions for 7 or 30 days and under aerobic conditions for 7 days. For colony forming units (CFU) assay, serial dilutions of cultures growing with or without diesel fuel as sole carbon source were plated in LB agar until colony development.
RNA extraction and RNA library preparation
Total RNA was isolated from P. extremaustralis cultures using Trizol method as described by Gomez-Lozano [15]. Cultures under aerobic or microaerophilic conditions were harvested after 24 h for RNA extraction. RNA quality was analyzed using an Agilent Bioanalyzer. rRNA depletion was performed using the MICROBExpress Kit (Ambion) with the addition of 5S oligonucleotides as was previously described [15]. The samples were validated with an Agilent 2100 Bioanalyzer (Agilent Technologies) after each step, and the final concentration was measured using a Qubit 2.0 Fluorometer (Invitrogen). Directional libraries were prepared with ScriptSeq v2RNA-Seq Library Preparation Kit (Epicentre) and were sequenced using the Illumina HiSeq 2000 platform with a paired-end protocol. Generated reads were of 100 nt lengths. For each condition duplicated independent RNA extraction and libraries were used.
RNA-seq data analysis
Data were de-multiplexed by Beckman Coulter Genomics. Reads alignment in P. extremaustralis genome and transcript abundance levels were quantified using the reads per kilobase per million mapped reads (RPKM) performed with the Rockhopper software [27] using default parameters. Differential gene expression was considered only with P < 0.05 and Q < 0.05. Spearman correlation analysis of normalized counts was performed to verify the concordance between replicates in aerobic and microaerophilic cultures (Fig. S1). Functional enrichment of differentially expressed genes was determined using Blast2GO software [10] by assigning the GO category to all genome sequences and to the differentially expressed genes. Additionally, in silico metabolic analysis was performed using Ipath2 software [43].
Quantitative real time PCR experiments (RT qPCR)
Total RNA of P. extremaustralis was extracted from E2 cultures supplemented with diesel fuel, diesel fuel + KNO3 or LB cultures supplemented with KNO3 under both aerobic and microaerophilic conditions using the Total RNA Extraction Kit (RBC Biosciences) as was described before [38]. After treatment with DNaseI, cDNA was obtained using random hexamers (Promega) and AMV retrotranscriptase following the manufacturer’s instructions. At least three independent cultures were analyzed for each condition. RT qPCR was performed using a LightCycler (DNA Engine M.J. Research) and Real Time PCR mix (EvaGreen qPCR Mix Plus, no Rox). alkB gene was analyzed using the following primers: 5′AACTACMTCGARCAYTACGG 3′ and 5′ TGAMGATGTGGTYRCTGTTCC 3′. Validation of RNA-seq results for selected genes was performed with the following primers: flgA 5′ACTGTTCAGGGATGTGGTGG′3 and 5′ GCTTCTCCGGGCATTTTCAC′3; azu ′5′GATCGACAAGAGCTGCAGGA′3 and 5′AGAAACCCGTAGTCCGTACCC ′3 and argC 5′CGCAAAGTCTTGGTGTGCC′3 and 5′TCCAGTGCTTCTGGAATGC′3. The 16S rRNA gene using primers 5′AGCTTGCTCCTTGATTCAGC′3 and 5′AAGGGCCATGATGACTTGAC′3 was used as reference for normalization of expression levels of target genes in each condition. The cycling conditions were as follows: denaturation at 95 °C for 5 min, 40 cycles at 95 °C for 25 s, 60 °C for 15 s, and 72 °C for 15 s, with the fluorescence acquisition at 80 °C in single mode. Relative changes in the expression of individual genes at aerobic and microaerophilic conditions in the different culture media tested were obtained through the relative standard curve method [20].
Hydrocarbon degradation
Seven-day cultures were analyzed to determine diesel fuel degradation. An uninoculated culture was used as a control and was considered as 100% of remnant hydrocarbon. Remnant hydrocarbons were extracted as described by Tribelli et al. [36]. Briefly, each culture was extracted with 20% v/v n-hexane and the solvent phase was dehydrated using anhydrous sodium sulphate. The samples were analyzed by GC using an Agilent 7820A gas chromatograph equipped with a Agilent HP-5 (30 m, 0.32 mm, 0.25 µm) column. The injector temperature was 250 °C, and the detector’s 280 °C. All samples were run at 60 °C for 10 min, then ramped to 155 °C at 5 °C min−1, 1 min at 155 °C then to 250 °C at 10 °C min−1 and then held for 50 min. For degradation assays, triplicate independent cultures were used. The percentage of residual hydrocarbon was calculated comparing the area of the GC chromatogram of the cultures with those of the control without bacteria. For alkane degradation, the area below each peak was compared. A constant peak with a RT of 26.5 min present in the YPF diesel fuel was use as internal control.
alkB bioinformatic analysis
To characterize the genomic region containing alkB, bioinformatic analysis were performed using tools included in the RAST server [4] and Pseudomonas.com site. alkB gene promoter region was analyzed using the Virtual Footprint tool available in PRODORIC Database [28]. Transcription binding sites, searched using 500 bp upstream ATG of alkB genes belonging to P. extremaustralis 14-3b (PE143B_0112385), P. aeruginosa PAO1 (PA1525 and PA2574 for alkB2 and alkB1, respectively), P. protegens Pf-5 (PFL_2935) and P. fluorescens SBW25 (PFLU3535), were compared.
Statistical analysis
The significance of the differences among different conditions was evaluated by the Mann–Whitney test with confidence levels at > 95% (i.e., P < 0.05 was considered as significant) [24]. Fisher’s test was used for gene over-representation analysis [13].
Data availability
RNA-seq data were deposited in the European Molecular Biology Laboratory under accession number E-MTAB-5440.
Results and discussion
Expression profile under microaerophilic conditions in P. extremaustralis
The RNA expression profile of P. extremaustralis cultures growing under microaerophilic or aerobic conditions in LB cultures supplemented with KNO3 revealed 5977 transcripts, including 187 putative regulatory RNAs. Rockhopper software analysis allowed the identification of genes differentially regulated at micro-aerobiosis (relative to aerobiosis) with statistical relevance. Under low oxygen conditions 357 genes were down-regulated and 337 were up-regulated (P < 0.05 and Q < 0.05, Table S1 and S2). This technique also revealed the existence of 53 novel intergenic sRNAs with differential expression.
To validate RNA-seq results a comparison with RT qPCR assay of some randomly selected genes (flgA, azu and argC) was performed. The genes flgA, azu and argC codify for the flagellar basal body P-ring biosynthesis protein FlgA, azurin and N-acetyl-gamma-glutamyl-phosphate reductase, respectively. While flgA and azu showed correlation between both analytical techniques (Table S3), in argC, both RNA-seq and RTqPCR assays presented low expression under microaerophilic conditions although the RT qPCR experiments showed significant differences (Table S3) In addition, RT qPCR experiments of other genes previously studied in P. extremaustralis under microaerophilic conditions was compared with the RNA-seq profile. These genes included phaC, encoding a polyhydroxybutyrate (PHB) synthase [39], and ccoN and cioA, both low oxygen affinity cytochromes [40] which were up-regulated, down-regulated or without changes, respectively, under microaerophilic conditions showing a concordance between both techniques.
To analyze RNA-seq results, differentially expressed genes were classified by functions (Fig. 1, Table S1 and S2). Genes involved in anaerobic metabolism (denitrification, arginine and pyruvate fermentation), transport and secretion, and different oxygenase and dioxygenase proteins as well as biological regulation were found to be up-regulated. Additionally, genes encoding regulatory proteins involved in low oxygen response, including narL, aer and bacterial chemotaxis related genes cheY, cheX, cheW and cheA were up-regulated as well. In contrast, genes involved in aminoacid metabolism, ethanol oxidation, regulator proteins and drug and metal resistance were down-regulated. To determine if some functions were over-represented in the up-regulated or the down-regulated differentially expressed gene data set, Blast2GO analysis was carried out. For down-regulated genes, amino acid metabolism and metal binding categories were over-represented (Fisher’s test using Blast2GO, P < 0.05, Fig. S2a). Blast2GO analysis of the up-regulated genes showed that genes encoding oxygen binding proteins, regulatory and signal transduction elements and kinase activity constituted the over-represented categories (Fig. S2b).
These results are aligned with those reported in P. aeruginosa growing under anaerobic conditions [40], where the activation of fermentation and alternative respiratory chain (denitrification) processes, together with the down regulation of the genes involved in translation and biosynthesis of aminoacids were observed.
Surprisingly, P. extremaustralis genes involved in n-alkane degradation were up-regulated under microaerophilic conditions (Table S1).
Genetic organization and expression of hydrocarbon degradation genes under microaerophilic conditions
Alkane degradation related genes were widely studied in P. putida Gpo1 and in P. aeruginosa PAO1 [1, 11, 21, 34]. Whereas P. putida Gpo1 presents the alkane degradation genes placed in a plasmid, P. aeruginosa PAO1 harbors in its chromosome two different alkane monooxygenase, one hydrocarbon degradation facilitator protein coding gene and other ubiquitous genes related to alkane degradation such as rubredoxine (alkG), rubredoxine reductase (alkT), aldehyde dehydrogenase and alcohol dehydrogenase and those belonging to β-oxidation pathway. In P. extremaustralis, genes involved in alkane degradation were located at different chromosomal positions. A putative operon composed by alkB and two genes encoding protein activators for alkane oxidation (praA and praB) were present in P. extremaustralis. This genetic organization was also found in P. protegens Pf-5, and P. fluorescens SBW25, two closely related species to P. extremaustralis (Fig. 2). alkS, a gene encoding a positive regulator of the alkBFGHJKL operon and present in the OCT plasmid in P. putida Gpo1 [6] was not found in P. extremaustralis’ genome. Surprisingly, P. extremaustralis genes involved in n-alkane degradation were expressed differently in microaerophilic conditions in LB cultures compared with aerobic cultures. The alkB gene, encoding the key enzyme for alkane degradation, alkane 1-monooxygenase, praA and praB encoding hydrocarbon facilitating proteins, and other genes related to this pathway, such as those encoding alcohol and aldehyde dehydrogenases, were up-regulated under low oxygen conditions (Fig. 3, Table S1). Additionally, genes involved in steps of fatty acid β-oxidation were also up-regulated, while rubredoxin coding genes (alkG and alkT) necessary for the oxidation reaction of alkanes were expressed similarly in aerobic and microaerophilic conditions (Fig. 3). A transcriptional study in P. aeruginosa PAO1, comparing the expression profile in jet fuel and glycerol supplemented cultures, showed that both alkB1 and alkB2 were induced in presence of jet fuel, although at different levels [16]. A quantitative proteomic study in this bacterium showed that AlkB2 was highly expressed in response to octadecane as carbon source in contrast to AlkB1, which was expressed equally in presence and absence of octadecane [22]. RT qPCR for alkB gene expression analysis was performed in minimum medium supplemented with diesel fuel as carbon source, with and without KNO3 under both aerobic and microaerophilic conditions. The latter resulted in an over-expression of the alkB gene compared with aerobic conditions (Fig. 4). Although a higher fold change was observed in KNO3 supplemented cultures, this difference resulted not significant (Fig. 4, Mann–Whitney test, P > 0.05). The promoter region of the alkB gene was analyzed in silico in P. extremaustralis and aligned with the corresponding regions from P. aeruginosa PAO1, P. protegens Pf-5 and P. fluorescens SWB25. A putative binding site for the anaerobic global regulator, Anr, was identified upstream of the alkB genes in P. extremaustralis and P. fluorescens SWB25 and of alkB1 in P. aeruginosa PAO1 (Fig. 2) suggesting that this microaerophilic degradation pathway could be extended to others alkane degraders Pseudomonas. Anr is a global regulatory protein that controls the transition from aerobiosis to micro-aerobiosis as well as the expression of genes involved in anaerobic metabolism [32, 35, 37, 38, 40, 41]. The presence of an Anr-box in the promoter region of the alkB gene was in line with both the higher expression under low oxygen conditions in P. extremaustralis, and the transcriptional data of P. aeruginosa PAO1 showing higher expression of alkB1 in hypoxia [2].
Alkane degradation and the ß-oxidation pathway in P.extremaustralis. Gray arrows and boxes represent genes without differences in their expression. Green arrows and boxes indicate up-regulated functions under low oxygen tension. OM outer membrane, IM inner membrane, AlkB alkane monooxygenase B, R rubredoxin, RRed rubredoxin reductase, ADH alcohol dehydrogenase, ALDH aldehyde dehydrogenase. Gene name and its respective locus tag in P.extremaustralis are indicated (color figure online)
Quantitative Real Time PCR: total RNA was extracted from P. extremaustralis cultures grown at microaerophilic and aerobic conditions with and without KNO3. alkB expression was normalized using rRNA 16S gene expression for each condition and a ratio between aerobic and microaerophilic condition was calculated. Values represent mean ± standard deviation (SD) of three independent experiments
Hydrocarbon degradation under low oxygen conditions
The up-regulation of the alkB gene observed in LB cultures without hydrocarbon under microaerophilic conditions, and the presence of an Anr-box like in the promoter zone of alkB, led to hypothesize that micro-aerobiosis was the preferred condition that allowed P. extremaustralis to grow using diesel fuel as the sole carbon source. Hydrocarbon degradation was analyzed in aerobic and microaerophilic cultures in minimal medium supplemented with diesel fuel as carbon source.
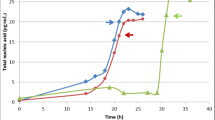
Pseudomonas extremaustralis was able to grow under microaerophilic conditions using diesel fuel as the sole carbon source, consuming 15.95 ± 2.97% of the diesel fuel after 7 days of growth. However, under aerobic conditions no hydrocarbon degradation was observed. These results are aligned with those previously observed in biofilm cultures [36]. The degradation under low oxygen conditions involved the C13–C19 aliphatic hydrocarbon fraction, including branched hydrocarbons as pristane, but not phytane (Fig. 5). To analyze the possibility that intra-aerobic hydroxylation during denitrification [8] could be used by P. extremaustralis, alkane degradation with and without the addition of KNO3 was tested. No differences in alkane degradation were observed under those conditions (Fig. 5).
To study the effect of diesel fuel degradation on P. extremaustralis growth, CFU/ml were calculated after 7 days in both aerobic and microaerophilic cultures, including control cultures without carbon source (Fig. 6). There was no difference in viable bacterial counts under aerobic conditions between cultures supplemented with diesel fuel and those without carbon source, being the result in agreement with the absence of alkane degradation in this condition. Under microaerophilic conditions, a slight increment in CFU/ml (Mann–Whitney test, P = 0.07) was observed with diesel fuel as carbon source in comparison with control cultures. The CFU/ml increment in diesel fuel supplemented microaerophilic cultures together with the low alkane degradation led to hypothesize that, the use of alkanes in microaerophilic conditions tends to support cellular viability more than bacterial replication. To test this hypothesis, a long-term culture (30 days) was analyzed under microaerophilic conditions showing again a small significant increase (Mann–Whitney test, P = 0.04) in CFU/ml in the diesel fuel supplemented media relative to the control without carbon source reaching a value of 2.6 × 106 ± 2.7 × 105 CFU/ml and 1.2 × 106 ± 2.6105 CFU/ml, respectively. As previously described [12], Pseudomonas species are able to use, under microaerophilic conditions, some metabolic pathways like pyruvate fermentation allowing survival for prolonged periods (18 days) without active growth. In this work not only an increase in P. extremaustralis survival but also a slow active growth was observed. Overall, the transcriptomic analysis allowed the identification of the unexpected over-expression under low oxygen conditions of necessary genes for hydrocarbon degradation in LB cultures. Our results showed that in P. extremaustralis, alkB expression, and alkane degradation are linked and dependent on the oxygen tension. The presence of an Anr-box like in the promoter region of the alkB gene suggests the microaerophilic induction of this gene. Interestingly, alkS the positive regulator of alkane degradation in P. putida Gpo1 [6, 17, 18], is absent in P. extremaustralis as well as in other Pseudomonas species such as P. aeruginosa PAO1 [34]. Additionally, it has been reported that in P. putida Gpo1 alkB expression is influenced by the level of expression of the cytochrome ubiquinol oxidase in addition to catabolic control [11]. The results showed not only a higher expression of alkB and other relevant genes but also diesel fuel degradation under conditions of oxygen limitation. Moreover, experiments performed in diesel fuel and glucose supplemented cultures showed similar alkane degradation (16.9 ± 3.8%) after 7 days in line with the hypothesis of other conditions rather than carbon source as the main regulatory factor in P. extremaustralis. The alkane degradation capability of P. extremaustralis, a bacterium isolated from a pristine environment, could be related to the fact that alkanes can be produced by different organisms from fatty acid metabolites, and found, for example, as components of plant cuticle waxes, insect pheromones and also in microorganisms such as cyanobacteria [33].
Interestingly, our results suggest that alkane degradation could be important for long-term survival and slow growth of this bacterium using diesel fuel as carbon source under microaerophilic conditions. Microaerophilic conditions could be found in a recent oil contaminated environment, where pollutants generate a strong impact in habitat quality affecting different environmental properties such as water flow, oxygen availability, nutrients, and light access [33]. As already described, when an oil spill occurs, a change in microbial communities’ composition begins [26]. In the Deepwater Horizon oil spill, aliphatic hydrocarbons degrading bacteria with large duplication times and more generalist strategies were the first ones to react to the environment change, being further replaced by most active degraders [29].
This study provides evidence of the activation of the alkane degradation pathway under microaerophilic conditions, regardless the presence of environmental hydrocarbon in a bacterium isolated from a pristine environment. This metabolism could represent an adaptive advantage in a changing environment by supporting survival under low oxygen conditions as found in recent oil contaminated sites. The role of this kind of microorganism should be considered as an important factor for the design of bioremediation strategies.
References
Alonso H, Kleifeld O, Yeheskel A, Ong PC, Liu YC, Stok JE, De Voss JJ, Roujeinikova A (2014) Structural and mechanistic insight into alkane hydroxylation by Pseudomonas putida AlkB. Biochem J 460:283–293. https://doi.org/10.1042/BJ20131648
Alvarez-Ortega C, Harwood CS (2007) Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65:153–165. https://doi.org/10.1111/j.1365-2958.2007.05772.x
Ayub ND, Tribelli PM, López NI (2009) Polyhydroxyalkanoates are essential for maintenance of redox state in the Antarctic bacterium Pseudomonas sp. 14-3 during low temperature adaptation. Extremophiles 13:59–66. https://doi.org/10.1007/s00792-008-0197-z
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21. https://doi.org/10.1007/s00253-006-0748-0
van Beilen JB, Panke S, Lucchini S, Franchini AG, Röthlisberger M, Witholt B (2001) Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621–1630. https://doi.org/10.1099/00221287-147-6-1621
Berthe-Corti L, Fetzner S (2002) Bacterial metabolism of n-alkanes and ammonia under oxic, suboxic and anoxic conditions. Acta Biotechnol 22:299–336
Callaghan AV (2013) Enzymes involved in the anaerobic oxidation of n-alkanes: from methane to long-chain paraffins. Front Microbiol 4:89
Chayabutra C, Ju LK (2000) Degradation of n-hexadecane and its metabolites by Pseudomonas aeruginosa under microaerobic and anaerobic denitrifying conditions. Appl Environ Microbiol 66:493–498. https://doi.org/10.1128/AEM.66.2.493-498.2000
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genom. https://doi.org/10.1155/2008/619832
Dinamarca MA, Aranda-Olmedo I, Puyet A, Rojo F (2003) Expression of the Pseudomonas putida OCT plasmid alkane degradation pathway is modulated by two different global control signals: evidence from continuous cultures. J Bacteriol 185:4772–4778. https://doi.org/10.1128/JB.185.16.4772-4778.2003
Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M (2004) Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. https://doi.org/10.1128/JB.186.14.4596-4604.2004
Fisher R (1934) Statistical methods for research workers. Oliver and Boyd, Edinburgh
Fonseca P, Moreno R, Rojo F (2011) Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ Microbiol Rep 3:329–339. https://doi.org/10.1111/j.1758-2229.2010.00229.x
Gómez-Lozano M, Marvig RL, Molin S, Long KS (2012) Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol 14:2006–2016. https://doi.org/10.1111/j.1462-2920.2012.02759.x
Gunasekera TS, Striebich RC, Mueller SS, Strobel EM, Ruiz ON (2013) Transcriptional profiling suggests that multiple metabolic adaptations are required for effective proliferation of Pseudomonas aeruginosa in jet fuel. Environ Sci Technol 47:13449–13458. https://doi.org/10.1021/es403163k
Hernández-Arranz S, Moreno R, Rojo F (2013) The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ Microbiol 15:227–241. https://doi.org/10.1111/j.1462-2920.2012.02863.x
Kok M, Oldenhuis R, Van Der Linden MPG, Raatjes P, Kingma J, Van Lelyveld PH, Witholt B (1989) The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J Biol Chem 264:5435–5441
Lageveen RG, Huisman GW, Preusting H, Ketelaar P, Eggink G, Witholt B (1988) Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol 54:2924–2932
Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinform 6:62. https://doi.org/10.1186/1471-2105-6-62
Liu H, Liang R, Tao F, Ma C, Liu Y, Liu X, Liu J (2012) Genome sequence of Pseudomonas aeruginosa strain SJTD-1, a bacterium capable of degrading long-chain alkanes and crude oil. J Bacteriol 194:4783–4784. https://doi.org/10.1128/JB.01061-12
Liu H, Sun W-B, Liang R-B, Huang L, Hou J-L, Liu J-H (2015) iTRAQ-based quantitative proteomic analysis of Pseudomonas aeruginosa SJTD-1: a global response to n-octadecane induced stress. J Proteom 123:14–28. https://doi.org/10.1016/j.jprot.2015.03.034
López NI, Pettinari MJ, Stackebrandt E, Tribelli PM, Põtter M, Steinbüchel A, Méndez BS (2009) Pseudomonas extremaustralis sp. nov., a poly(3-hydroxybutyrate) producer isolated from an antarctic environment. Curr Microbiol 59:514–519. https://doi.org/10.1007/s00284-009-9469-9
Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18(1):50–60
Martínez-Antonio A, Collado-Vides J (2003) Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol 6:482–489. https://doi.org/10.1016/j.mib.2003.09.002
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, Han J, Holman HY, Hultman J, Lamendella R, Mackelprang R, Malfatti S, Tom LM, Tringe SG, Woyke T, Zhou J, Rubin EM, Jansson JK (2012) Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to deepwater horizon oil spill. ISME J 6:1715–1727. https://doi.org/10.1038/ismej.2012.59
McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B (2013) Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. https://doi.org/10.1093/nar/gkt444
Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D (2005) Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. https://doi.org/10.1093/bioinformatics/bti635
Rodriguez-R LM, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT (2015) Microbial community successional patterns in beach sands impacted by the deepwater horizon oil spill. ISME J 9:1928–1940. https://doi.org/10.1038/ismej.2015.5
Rojo F (2005) Specificity at the end of the tunnel: understanding substrate length discrimination by the AlkB Alkane hydroxylase. J Bacteriol 187:19–22. https://doi.org/10.1128/JB.187.1.19-22.2005
Rojo F (2009) Degradation of alkanes by bacteria: minireview. Environ Microbiol 11:2477–2490. https://doi.org/10.1111/j.1462-2920.2009.01948.x
Sawers RG (1991) Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol 5:1469–1481
Schirmer A, Rude MA, Li X, Popova E, Del Cardayre SB (2010) Microbial biosynthesis of alkanes. Science 329:559–562. https://doi.org/10.1126/science.1187936
Smits THM, Witholt B, van Beilen JB (2003) Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 84:193–200. https://doi.org/10.1023/a:1026000622765
Tribelli PM, Hay AG, López NI (2013) The global anaerobic regulator Anr, is involved in cell attachment and aggregation influencing the first stages of biofilm development in Pseudomonas extremaustralis. PLoS One 8:e76685. https://doi.org/10.1371/journal.pone.0076685
Tribelli PM, Di Martino C, López NI, Raiger Iustman LJ (2012) Biofilm lifestyle enhances diesel bioremediation and biosurfactant production in the Antarctic polyhydroxyalkanoate producer Pseudomonas extremaustralis. Biodegradation 23:645–651. https://doi.org/10.1007/s10532-012-9540-2
Tribelli PM, Méndez BS, López NI (2010) Oxygen-sensitive global regulator, anr, is involved in the biosynthesis of poly(3-Hydroxybutyrate) in Pseudomonas extremaustralis. J Mol Microbiol Biotechnol 19:180–188. https://doi.org/10.1159/000320261
Tribelli PM, Nikel PI, Oppezzo OJ, López NI (2013) Anr, the anaerobic global regulator, modulates the redox state and oxidative stress resistance in Pseudomonas extremaustralis. Microbiology 159:259–268. https://doi.org/10.1099/mic.0.061085-0
Tribelli PM, Venero ECS, Ricardi MM, Gómez-Lozano M, Raiger Iustman LJ, Molin S, López NI (2015) Novel essential role of ethanol oxidation genes at low temperature revealed by transcriptome analysis in the antarctic bacterium Pseudomonas extremaustralis. PLoS One 10:e0145353. https://doi.org/10.1371/journal.pone.0145353
Trunk K, Benkert B, Quäck N, Münch R, Scheer M, Garbe J, Jänsch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D (2010) Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. https://doi.org/10.1111/j.1462-2920.2010.02252.x
Ugidos A, Morales G, Rial E, Williams HD, Rojo F (2008) The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ Microbiol 10:1690–1702. https://doi.org/10.1111/j.1462-2920.2008.01586.x
Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB, Throne-Holst M (2007) Bacterial metabolism of long-chain n-alkanes. Appl Microbiol Biotechnol 76:1209–1221. https://doi.org/10.1007/s00253-007-1119-1
Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P (2011) IPath2.0: interactive pathway explorer. Nucleic Acids Res 39:W412–W415. https://doi.org/10.1093/nar/gkr313
Acknowledgements
This work was partially supported by grants from UBA, CONICET, and ANPCyT. PMT, MMR, LJRI and NIL are career investigators from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). RNA-seq experiments were performed by PMT at Dr. Molin’s Lab supported by a short term EMBO fellowship. Authors want to thank to anonymous reviewers for the helpful criticisms.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tribelli, P.M., Rossi, L., Ricardi, M.M. et al. Microaerophilic alkane degradation in Pseudomonas extremaustralis: a transcriptomic and physiological approach. J Ind Microbiol Biotechnol 45, 15–23 (2018). https://doi.org/10.1007/s10295-017-1987-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1987-z