Abstract
Poly(β-L-malic acid) (PMA) is a natural biopolyester that has pharmaceutical applications and other potential uses. In this study, we examined PMA production by 56 strains of the fungus Aureobasidium pullulans representing genetically diverse phylogenetic clades. Thirty-six strains were isolated from various locations in Iceland and Thailand. All strains from Iceland belonged to a newly recognized clade 13, while strains from Thailand were distributed among 8 other clades, including a novel clade 14. Thirty of these isolates, along with 26 previously described strains, were examined for PMA production in medium containing 5% glucose. Most strains produced at least 4 g PMA/L, and several strains in clades 9, 11, and 13 made 9–11 g PMA/L. Strains also produced both pullulan and heavy oil, but PMA isolated by differential precipitation in ethanol exhibited up to 72% purity with no more than 12% contamination by pullulan. The molecular weight of PMA from A. pullulans ranged from 5.1 to 7.9 kDa. Results indicate that certain genetic groups of A. pullulans are promising for the production of PMA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biopolymers have the potential to replace petroleum-derived materials in numerous applications as bioplastics, biocomposites, adhesives, films or fibers. First-generation bioplastics include the familiar biopolymers poly(β-lactic acid) (PLA) and poly(3-hydroxyalkanoates) (PHA). Although a variety of diverse microbial polyesters is known, research is needed to discover and develop promising second-generation biomaterials [12, 20]. One such biopolymer of interest is poly(β-malic acid) (PMA). Because it is biocompatible, degradable, and water soluble, PMA has primarily been studied for biomedical uses as a drug carrier [1, 11]. However, water-insoluble PMA derivatives have been used to make various solid devices, compression molded pellets, films, microparticles and nanoparticles, although to date none commercially [23].
PMA was first identified by Shimada et al. [18] as a protease inhibitor from Penicillium cyclopium and later from the myxomycete (slime mold) Physarum polycephalum [3, 5], where it was determined to be a DNA polymerase inhibitor. P. polycephalum currently is used to produce PMA for pharmaceutical applications [11]. Nagata et al. [15] reported that isolates of the fungus Aureobasidium also produce PMA in high yields. In a small screening study, Liu and Steinbuchel [10] reported that 4 of 8 strains of Aureobasidium produced up to 1.1 g PMA/L prior to optimization. These authors suggested that if PMA could be produced more economically, it might find applications in the production of detergents, biodegradable plastics, or other biomaterials.
Aureobasidium pullulans is a polymorphic fungus, considered to be a filamentous ascomycete in class Dothideomycetes, subclass Dothideomycetidae [4, 17]. A. pullulans is well known as the source of the commercial polysaccharide, pullulan [8, 19]. The fungus also produces numerous other valuable bioproducts, including industrial enzymes, PMA, and a novel heavy oil [2, 7, 15]. We recently completed a multilocus molecular phylogeny of A. pullulans [13]. Interestingly, certain phylogenetically defined clades produced high levels of specific bioproducts, including pullulan, xylanase, and heavy oil [13, 14]. The objective of this study is to examine PMA production by genetically diverse phylogenetic clades of A. pullulans, in an effort to identity groups that produce high levels of PMA.
Materials and methods
A. pullulans isolates and classification
Thirty-six strains were isolated for this study between October 2009 and February 2010, designated RSU strains (Table 1). RSU strains 1–8 were collected from Iceland, with the remaining RSU strains collected from various provinces in Thailand. The method of isolation and phylogenetic analysis was described by Manitchotpisit et al. [13]. All isolates were classified using morphological characteristics and DNA sequence analyses of two loci, ITS and β-tubulin (BT2). All sequences were determined from bidirectional sequencing and aligned with our previous sequence data using ClustalW [22]. Aureobasidium sp. strain CU 30 was chosen as an outgroup species based on our previous studies [13]. Maximum parsimony analysis was performed using PAUP* [21] version 4.0b10. DNA sequences determined in this study were deposited in GenBank under accession numbers JF419588-JF419659, JF682345-JF682347, FJ150905.1, FJ150906.1, FJ157867.1, and FJ157869.1. Other strains used in this study were previously described [13] or obtained from the ARS Culture Collection, Peoria, IL (NRRL strains) or the Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands (CBS strains).
PMA production by strains of A. pullulans
A screen of PMA production by 56 strains of A. pullulans was conducted by culturing each isolate in 10 mL of modified pullulan production medium [13] containing 5% (w/v) glucose, 0.06% (w/v) peptone, 0.04% (w/v) yeast extract, 0.5% (w/v) K2HPO4, 0.04% (w/v) MgSO4.7H2O, and 0.1% (w/v) NaCl, pH 6.5, in 50 mL flasks cultured for 7 days at 25°C with shaking at 200 rpm. Cells were removed by centrifugation at 8,000 rpm (10,000 × g) for 20 min. Culture supernatants were analyzed for PMA and pullulan by analyzing malic acid and glucose yields, respectively, before and after acid hydrolysis. Samples (500 μL) of each supernatant were mixed with an equal volume of 1.0 M H2SO4 and hydrolyzed by incubation at 90°C for 9 h. Hydrolyzed samples and unhydrolyzed controls were diluted 1:5 in an HPLC dilution buffer (19 mM nitric acid and 34 mM propionic acid, which served as a microbiocide and internal standard, respectively) and applied to an Aminex HPX-87H column (Bio-Rad, Hercules, CA) equilibrated with 15 mM nitric acid at a flow rate of 0.5 mL/min at 60°C. Eluate from the column was analyzed by an RI detector. Increases in free malic acid and glucose in hydrolyzed samples were interpreted as representing PMA and pullulan, respectively. Pullulan (Showa Denko, Tokyo, Japan) and purified PMA (the kind gift of Dr. Eggehard Holler) from P. polycephalum (M w 30,000) served as controls. The experiments were performed in triplicate and standard errors are reported.
Comparison of PMA, pullulan, and heavy oil production
To study potential relationships among PMA, pullulan and heavy oil production, representative strains were grown for 7 days in 50 mL of modified pullulan production medium in 250 mL flasks. The pH and OD600 of each culture were recorded, and cultures were centrifuged at 8,000 rpm (10,000 × g) for 20 min to prepare supernatants. Samples (1 mL) were retained for PMA and pullulan determinations using HPLC as described above. The remaining culture supernatants were adjusted to pH 5.0 by the dropwise addition of 30% (w/v) CaCO3. Pullulan was precipitated by the addition of one volume of 95% ethanol followed by centrifugation at 10,000 rpm (16,900 × g) for 30 min. PMA was then precipitated by the addition of an additional volume of 95% ethanol, followed by incubation at 4°C overnight and centrifugation at 10,000 rpm (16,900 × g) for 30 min. Precipitated PMA was dissolved in 200 mL of Milli-Q water (Millipore, Billerica, MA) and lyophilized. Heavy oil was extracted from the cell pellets using methyl ethyl ketone as described by Manitchotpisit et al. [14]. The experiments were performed in triplicate and standard errors are reported.
Characterization of isolated PMA
Lyophilized PMA samples were dissolved in Milli-Q water at 1.0% (w/v) and assayed for PMA content by HPLC as described above. For analysis by nuclear magnetic resonance spectroscopy (NMR), samples were dissolved in D2O to a final concentration of 2 mg/mL and analyzed by 1H-NMR using a Bruker Avance 500 spectrometer (Bruker Biospin Corp., Billerica, MA) equipped with a 5 mm broadband inverse (BBI) probe with z-gradient. NMR data were processed with Topspin v1.3 software and the chemical shifts were reported as parts per million from external tetramethylsilane (TMS) based on the lock signal. Pullulan (Showa Denko) and purified PMA from P. polycephalum (M w 30,000) served as NMR standards. Molecular weight distributions were analyzed by size exclusion chromatography, using a Shodex SB-806 M HQ column (Showa Denko, Tokyo, Japan) eluted with 0.05 M sodium nitrate at room temperature and a flow rate of 0.5 mL/min. Separations were monitored using a Shodex OR-1 optical rotation detector (Showa Denko K.K.). Pullulan standards of varying known sizes (Showa Denko) were used to estimate the molecular weight of PMA.
Results and discussion
Isolation and classification of A. pullulans strains
Thirty-six strains of A. pullulans were isolated for this study, designated as RSU strains (Table 1). RSU strains 1–8 were collected in Iceland, and RSU strains 9–36 were collected from various provinces in Thailand, between October, 2009 and February, 2010. Most strains were isolated from leaf samples, consistent with the reputation of A. pullulans as a common epiphyte. All new isolates were deposited in the ARS Culture Collection, National Center for Agricultural Utilization Research, USDA, Peoria, IL, USA (accession numbers are shown in Table 1). New strains were classified using morphological characteristics and by DNA sequence analyses of the ITS and BT2 loci.
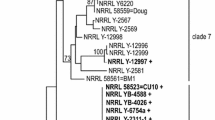
Phylogenetic analysis of the ITS region is useful to distinguish A. pullulans from other species [13], and by this measure all 36 new strains were confirmed as A. pullulans. However, ITS analysis cannot be used to differentiate individual isolates. On the other hand, BT2 sequences are useful to classify A. pullulans isolates into phylogenetic groups [13]. The BT2 region is convenient for this analysis because it involves relatively short PCR products (approximately 450 bp) and only one pair of primers is required [13]. Most of these isolates were classified into distinct phylogenetic clades, although strains RSU 10 and RSU 35 were assigned to clade 1 or 2. Fig. 1 shows a maximum parsimony tree of BT2 sequences of 63 A. pullulans strains, including the new isolates. The clade numbering system shown is based on our previous multilocus phylogenetic study [13]. Eight strains isolated from Iceland (RSU 1–8) all belong to newly recognized clade 13, which contains only strains from temperate climates. Clade 13 includes CBS strain 584.75, which is the exneotype strain for A. pullulans var. pullulans, as well as CBS strain 100,524, now considered A. pullulans var. pullulans [24]. Also included in clade 13 is CBS strain 591.75, identified as the best PMA producer among 8 strains examined by Liu and Steinbuchel [10]. Two new isolates from Thailand, strains RSU 11 and RSU 15, belong to a novel clade 14 (Table 1). The remaining 26 new isolates from Thailand were distributed among 8 previously described phylogenetic clades (clades 1 and 2, 2 isolates; clade 3, 11 isolates; clade 5, 1 isolate; clade 7, 3 isolates; clade 8, 2 isolates; clade 9, 4 isolates; and clade 11, 3 isolates). The variety of these strains supports our previous conclusion that Thailand is a source of genetically diverse isolates of A. pullulans [13].
Maximum parsimony tree of Aureobasidium pullulans isolates calculated with PAUP*. The bootstrap proportion from 1,000 repetitions appears above nodes. The clade numbering system is from the multilocus phylogenetic study of Manitchotpisit et al. [13]
Phenotypic observations of the new isolates were consistent with previous descriptions of A. pullulans [13]. Colonial morphologies grown on yeast malt agar for 7 days generally were smooth, moist, yeast-like, and pale pink. Strain RSU 12 exhibited a dark vinaceous pigment characteristic of clade 5. Isolates in clade 8 (RSU 20 and RSU 32) exhibited characteristic color rings of pale pink and orange. On malt extract agar plates, colonies generally consisted of white hyphae with dark olivaceous centers, except for isolates in clades 5, 8, and 11, which showed vinaceous, yellow, and white pigments, respectively. Microscopically, early cultures showed typical polymorphic forms of A. pullulans with blastospores, swollen cells, and pseudohyphae, with later cultures showing hyphae and chlamydospores.
PMA production by diverse phylogenetic clades of A. pullulans
Fifty-six strains of A. pullulans were examined for PMA production, including 30 of the new isolates described above and an additional 26 strains representing diverse clades of A. pullulans (Table 2). In preliminary studies, we tested the mineral salts medium used by Nagata et al. [15] and Liu and Steinbuchel [10]. However, we obtained higher yields of PMA from a modified pullulan production medium, subsequently used in these studies. Most strains produced PMA on this medium, and several strains in clades 9, 11, and 13 made 9–11 g PMA/L (Table 2). Clade 13 strain CBS 591.75, previously described as the best producer among eight strains of A. pullulans [10], produced 8.8 g PMA/L under these conditions.
Comparison of PMA, pullulan, and heavy oil production
PMA, pullulan, and heavy oil production were compared from strains associated with nine different clades (Table 3). Cultures were grown on a slightly larger scale (50 mL) to provide greater amounts of material for analysis. Growth yields varied considerably and were not obviously related to PMA yields. PMA yields also were somewhat different from those observed in smaller cultures, presumably due to the effects of scale. Nevertheless, strain RSU 7 from clade 13 was the best producer of PMA, consistent with results from the screening study. Only strain RSU 15 failed to produce PMA under these conditions, consistent with observations of low production by members of clade 14.
All strains produced both pullulan and heavy oil (Table 3). Strain NRRL Y-12974 was previously described as a high-level producer of pullulan [13], and new isolates RSU 7 and RSU 15 produced nearly 10 g pullulan/L. Strains CU 44 and CU 47 produced somewhat less pullulan than previously reported [13], possibly because of differences in growth and assay conditions. Strain CU 47 (clade 11) gave the highest yields of heavy oil. In most cases, the color of the oil produced corresponded to the color of the culture (Table 3). No relationships were apparent among the yields of PMA, pullulan, and oil. However, in preliminary experiments we observed that the addition of 3.0% (w/v) CaCO3 to growing cultures increased yields of PMA by 25–80% and decreased yields of both pullulan and oil (data not shown). Liu and Steinbuchel [10] cultured A. pullulans in medium without CaCO3, but added CaCO3 to culture supernatants to facilitate precipitation of Ca-PMA. On the other hand, Nagata et al. [15] included CaCO3 in the culture medium as a neutralizing agent, and Kurosawa et al. [6] later observed that the absence of CaCO3 favored the accumulation of heavy oil instead of PMA. Although culture pH is stabilized by the addition of CaCO3, it is not clear that pH is the sole factor influencing PMA production. Lee et al. [9] concluded that, in the slime mold P. polycephalum, CaCO3 can serve as a significant carbon source for PMA biosynthesis.
Characterization of isolated PMA
PMA was isolated by differential precipitation in ethanol, which preferentially separates PMA from pullulan. The PMA and pullulan content was assayed as described above (Table 4). Strains CU 22, CU 44, and CU 47 exhibited PMA of about 70% purity, with no more than 8% pullulan. Despite the fact that strain NRRL Y-12974 produced a great deal of pullulan, precipitated PMA from this strain showed 62% purity with 12% pullulan. This suggests that differential precipitation is relatively successful at separating pullulan from PMA.
Isolated PMA also was characterized by 1H-NMR spectroscopy. Fig. 2 shows spectra obtained for strains NRRL Y-12974 and CU 22, along with purified pullulan and PMA standards. PMA is characterized by multiplets due to methylene (-CH 2 -, peak b) and methine (-CH-, peak d) protons (panel D). Pullulan is characterized by peaks for the branched and unbranched alpha-1,4-linked (peak a) and the alpha-1,6-linked (peak c) glucosyl residues (panel C). Other pullulan signals at 3–4 ppm are due to the carbohydrate ring protons. Panel B of Fig. 2 shows that the preparation from strain NRRL Y-12974 contains PMA and likely some contaminating pullulan. Panel A shows the presence of PMA in the preparation from strain CU 22, and no contaminating pullulan is evident. However, the preparation may contain non-pullulan carbohydrate, as shown by anomeric proton peaks at 4.6 ppm and 5.1 ppm, and ring protons at 3.0-4.0 ppm. NMR spectra of strains CU 43, CU 44, CU 47, and RSU 7 appeared identical to that of CU 22 (data not shown).
We also examined the molecular weight of PMA produced by strains of A. pullulans. Nakajima-Kambe et al. [16] reported that the molecular weight of PMA from strain A-91 of Aureobasidium was 6-11 kDa. Similarly, Liu and Steinbuchel [10] reported a molecular weight of 3–5 kDa for PMA from A. pullulans, while PMA from the slime mold P. polycephalum is on the order of 50 kDa [11]. Consistent with these reports, we found that PMA from A. pullulans strains ranged from 5.1 to 7.9 kDa, while a PMA standard from P. polycephalum was 27 kDa (Table 4). Thus, isolated PMA preparations characterized in this study were all of relatively low molecular weight. It is possible that examination of additional strains will provide exceptions to this rule. In the case of the polysaccharide pullulan, it is clear that molecular weight varies greatly on a strain-specific basis [8].
In conclusion, most strains of A. pullulans in genetically diverse phylogenetic clades produced PMA, and certain clades included strains that produced relatively high levels of PMA (9–11 g/L). PMA isolated by differential precipitation in ethanol exhibited up to 72% purity with no more than 12% contamination by pullulan. The molecular weight of PMA from A. pullulans ranged from 5.1 to 7.9 kDa. Thus, certain genetic groups of A. pullulans are promising for the production of PMA.
References
Braud C, Bunel C, Vert M (1985) Poly(β-malic acid): a new polymeric drug-carrier. Evidence for degradation in vitro. Polym Bull 13:293–299
Deshpande MS, Rale VB, Lynch JM (1992) Aureobasidium pullulans in applied microbiology: a status report. Enzyme Microbial Technol 14:514–527
Fischer H, Erdmann S, Holler E (1989) An unusual polyanion from Physarum polycephalum that inhibits homologous DNA polymerase in vitro. Biochem 28:5219–5226
Hibbett DS et al (2007) A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547
Holler E, Angerer B, Achhammer G, Miller S, Windisch C (1992) Biological and biosynthetic properties of poly-L-malate. FEMS Microbiol Rev 103:109–118
Kurosawa T, Sakai K, Nakahara T, Oshima Y, Tabuchi T (1994) Extracellular accumulation of the polyol lipids, 3, 5-dihydroxydecanoyl and 5-hydroxy-2-decenoyl esters of arabitol and mannitol, by Aureobasidium sp. Biosci Biotech Biochem 58:2057–2060
Leathers TD (1989) Purification and properties of xylanase from Aureobasidium. J Ind Microbiol 4:341–348
Leathers TD (2002) Pullulan. In: Vandamme EJ, De Baets S, Steinbuchel A (eds) Biopolymers, vol 6, polysaccharides II: polysaccharides from eukaryotes. Wiley-VCH, Weinheim, pp 1–35
Lee BS, Maurer T, Kalbitzer HR, Holler E (1999) β-Poly(L-malate) production by Physarum polycephalum: 13C Nuclear magnetic resonance studies. Appl Microbiol Biotechnol 52:415–420
Liu S, Steinbuchel A (1996) Investigation of poly(β-L-malic acid) production by strains of Aureobasidium pullulans. Appl Microbiol Biotechnol 46:273–278
Ljubimova JY, Fujita M, Khazenzon NM, Lee BS, Wachsmann-Hogiu S, Farkas DL, Black KL, Holler E (2008) Nanoconjugate based on polymalic acid for tumor targeting. Chemico-Biol Interact 171:195–203
Luengo JM, Garcia B, Sandoval A, Naharro G, Olivera ER (2003) Bioplastics from microorganisms. Curr Opin Microbiol 6:251–260
Manitchotpisit P, Leathers TD, Peterson SW, Kurtzman CP, Li X-L, Eveleigh DE, Lotrakul P, Prasongsuk S, Dunlap CA, Vermillion KE, Punnapayak H (2009) Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aureobasidium pullulans. Mycol Res 113:1107–1120
Manitchotpisit P, Price NPJ, Leathers TD, Punnapayak H (2011) Heavy oils produced by Aureobasidium pullulans. Biotechnol Lett 33:1151–1157
Nagata N, Nakahara T, Tahuchi T (1993) Fermentative production of poly(β-L-malic acid), a polyelectronic biopolyester by Aureobasidium sp. Biosci Biotech Biochem 57:638–642
Nakajima-Kambe T, Hirotani N, Nakahara T (1996) Poly(beta-malic acid) production by the non-growing cells of Aureobasidium sp strain A-91. J Ferm Bioeng 82:411–413
Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98:1041–1052
Shimada K, Matsushi K, Fukumoto J, Yamamoto T (1969) Poly-(L)-malic acid—a new protease inhibitor from Penicillium cyclopium. Biochem Biophy Res Commun 35:619–624
Singh RS, Saini GK, Kennedy JF (2008) Pullulan: microbial sources, production and applications. Carbohydr Polym 73:515–531
Steinbuchel A, Valentin HE (1995) Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett 128:219–228
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associates, Sunderland
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vert M (1998) Chemical routes to poly(β-malic acid) and potential applications of this water-soluble bioresorbable poly(β-hydroxy alkanoate). Polym Degrad Stab 59:169–175
Zalar P, Gostincar C, de Hoog GS, Ursic V, Sudhadham M, Gunde-Cimerman N (2008) Redefinition of Aureobasidium pullulans and its varieties. Stud Mycol 61:21–38
Acknowledgments
The authors acknowledge RSU grant number 37/52 from the Research Center of Rangsit University for partial financial support. Appreciation is expressed for the kind assistance provided by Kristina Glenzinski, Melinda Nunnally, and Trina Hartman. Poly(β-L-malic acid) from P. polycephalum (M w 30,000) was the kind gift of Dr. Eggehard Holler, Cedars-Sinai Medical Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
“Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may be suitable. USDA is an equal opportunity provider and employer.”
Rights and permissions
About this article
Cite this article
Manitchotpisit, P., Skory, C.D., Peterson, S.W. et al. Poly(β-L-malic acid) production by diverse phylogenetic clades of Aureobasidium pullulans . J Ind Microbiol Biotechnol 39, 125–132 (2012). https://doi.org/10.1007/s10295-011-1007-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-1007-7