Abstract
To better engineer and analyze beneficial biofilms as well as to develop strategies to control detrimental biofilms (e.g., biomedical device-based infections), it is critical to quantify bacterial species compositions within biofilms. A non-invasive method is described here that determines local and overall bacterial concentrations within a biofilm, using optical microscopy and digital image analysis techniques. The method is based upon a calibration of cell fluorescence to known cell number concentrations and is verified by direct cell counts of destructive samples of cultivated biofilms. Two GFP mutants, each with unique emission colors were used with both epi-fluorescent microscopy and one-photon confocal microscopy to determine local spatial biofilm cell concentrations in pure and mixed-strain biofilms. Our microbial system comprises Pseudomonas putida containing either green fluorescent protein (GFP) or containing the red fluorescent protein (DsRed). Strains expressing a green or red fluorescent protein were detected by two different microscopy methods: epi-fluorescence and single-photon confocal laser scanning microscopy. Overall biofilm cell concentrations determined directly from destructive samples were in good agreement with non-invasive measurements of adherent cell concentrations calculated from the measured “integrated fluorescent density” minus any background fluorescence. Results show the areal cell concentration (cell number/area) determined from non-destructive direct counts in a pure culture or binary-strain biofilm varied with the biofilm depth. Use of this method to estimate local dynamic plasmid segregational loss and plasmid conjugation transfer kinetics will be reported in a subsequent manuscript.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial cells (predominantly bacteria) and their extracellular polymers associated with a substratum are called biofilms [3, 5, 6, 11]. Beneficial examples of biofilm processes can be found in wastewater treatment reactors, specialty commodity production, and xenobiotic waste degradation. However, biofilms can also create serious problems by causing significant increases in both frictional and heat-transfer resistances; by mediating material deterioration; by contaminating artificial organs, catheters, and endoprostheses; and by promoting serious, perhaps fatal, infections (e.g., cystic fibrosis). Biofilms are medically important since few diseases are caused by microbes that are planktonic; that is, non-adherent and free-floating. Bacteria bound within biofilms are remarkably difficult to treat with antibiotics or antimicrobials. The reasons for this are not clear, but recent work has provided some insights [5, 8, 27]. Antimicrobials may be readily degraded by specific enzymes secreted by the adherent bacteria or the chemical agent may fail to penetrate completely into the biofilm due to mass transfer/reaction rate limitations [26, 27]. In addition to combined mass transfer-reaction rate limitations, bacteria within biofilms may be less susceptible to antimicrobial compounds through a variety of metabolic, enzymatic, and genetic defense mechanisms that are not manifested by their planktonic counterparts. Biofilm-bound bacteria may enhance the segregational stability, expression, and rate of conjugative transfer of plasmid-DNA (often containing antibiotic resistance genes) between bacteria [2, 13–16]. This is important since antibiotic-resistant bacteria, in a biofilm, may transfer these resistance genes more readily to neighboring bacteria [2]. Gene transfer could also convert a previous avirulent commensal organism into a highly virulent pathogen or impart the phenotypic ability to tolerate or metabolize a xenobiotic compound.
Biofilms form on a wide range of inert and living surfaces and are associated with a number of significant beneficial (e.g., wastewater treatment) and damaging (e.g., fouling, corrosion, nosocomial infections, biomaterials infections and dental disease) processes [4]. Consequently, there is considerable interest in developing methods to study the architecture of, and metabolism within, such biofilms. Imaging of biofilms began with van Leeuwenhoek’s observations of dental plaque in the 17th century. The development of scanning electron microscopy (SEM) in the last 30 years has allowed imaging of the detailed structures of a wide range of biofilms [24]. Unlike SEM, other microscopic techniques, in particular, confocal laser scanning microscopy (CLSM), now allows the study of hydrated, viable biofilms. CLSM coupled with fluorescent and immunofluorescent stain combinations [14] have dramatically improved the ability to non-invasively dissect a bacterial biofilm. Such studies have shown biofilms to have a more open architecture [20], in which towers of biofilm mass were shown to be interspersed with water channels. Coupled with the development of various fluorescent stains, the CLSM can provide observations of a variety of cellular and biofilm community processes (e.g., multiple species enumeration, plasmid presence and transfer, bacterial cell viability, and specific gene expression via in situ 16s rRNA hybridization).
The green fluorescent protein (GFP) has emerged as a versatile reporter gene and in situ cell marker. Advantages such as species independence and the lack of a requirement for a promoter make GFP unique as a reporter gene [7]. GFP has become an especially valuable marker for nondestructively visualizing cells, particularly in biofilms. GFP in combination with confocal laser scanning microscopy (CLSM) has led to new insights into biofilm structural development and biological processes [12]. Several GFP variants with excitation and emission properties different from those of the wild-type protein have been developed [1]. One such variant, GFPuv [11] (Clontech, Palo Alto, CA), emits bright green light (maximum at 509 nm) or blue light when exposed to UV at 395 or 470 nm. Mutant proteins GFPmut2 and GFPmut3 [9] have emission maxima of 507 and 511 nm when excited by blue light (481 and 501 nm, respectively). Unlike GFPuv, GFPmut2 and GFPmut3 are not excited by UV light; a difference that allows differential imaging of these proteins in the same sample. A multiple-labeling technique based on two GFPs has been used for epifluorescent microscopy [21] and for one-photon confocal microscopy [10] for (a) mixed cultures of cells, where one species contained GFPuv and another species contained GFPmut2 or GFPmut3, and (b) a single species containing both GFPuv and GFPmut2 in the same cell. In all cases, these studies are qualitative, in that location and presence of the various GFP-expressing species (strains) are observed but exact absolute local concentrations of each microorganism were not determined.
This work reports the development of a non-invasive method to quantify local and overall bacterial concentrations within a biofilm. The method is based upon a calibration of cell fluorescence to known cell numbers concentrations and verified by direct cell counts of destructive samples of cultivated biofilms.
Materials and methods
Bacterial strains and media
Pseudomonas putida (Ppu) KT2440 strains were used throughout this entire study. Several of these strains, which are listed in Table 1, were kindly provided by Prof. Dr. Søren Molin (Technical University Denmark). All host strains Ppu KT2440 contain the mini-Tn7 transposon system expressing different mutant variants of the green fluorescent protein (GFP), using the Escherichia coli lac promoter, PA1/04/03. Gentamicin-resistant genes are also present in all of these strains [19]. The remaining strains were donated courtesy of Dr. Barth Smets (Technical University Denmark). Two of these Pseudomonas putida KT2440 stains express two different types of marker proteins, GFP and DsRed, respectively. LB Broth (Bacto tryptone 10 g/l, Bacto yeast extract 5 g/l, NaCl 4 g/l) and chemically defined medium (Na3C6H5O7, 129 mg/l; (NH4)2SO4, 2 g/l; Na2HPO4·2H2O, 6 g/l; KH2PO4, 3 g/l; NaCl, 3 g/l; MgCl2, 93 mg/l; CaCl2; 11 mg/l; Trace metal solution, 1 ml/l) supplemented with Gentamicin (10 μg/ml) were used for batch suspended cell cultures. The trace metal solution is composed of CaSO4·2H2O, 200 mg/l; FeSO4·7H2O, 200 mg/l; MnSO4·H2O, 20 mg/l; CuSO4·5H2O; 20 mg/l; ZnSO4·7H2O; 10 mg/l; CoSO4·7H2O, 10 mg/l; Na2MoO4·H2O; 10 mg/l; H3BO3, 5 mg/l [17]. Both GFP and DsRed are very stable in both complicated and chemically defined medium. For suspended culture inoculum, a sample of one colony from the streak plate was collected by sterile loop and added to 25 ml of 10 g/l LB Broth, then incubated at 30°C overnight. Bacterial strains were maintained in the 15% glycerol stock safely for an indefinite period of time at −80°C.
Biofilm reactor systems
A stainless-steel flow cell (Protofab, Bozeman, MT) was used for biofilm cultivation and non-invasive microscopic analyses (Fig. 1). The flow cell consists of a single flow-through channel (38 mm long, 12 mm wide, 2.5 mm deep). Glass coverslips (Erie Scientific) (48 mm wide × 65 mm long × 0.13–0.17 mm thick) were used to form the top and bottom of the flow channel. The entire system, except the flow cells, was sterilized by autoclaving. To sterilize the stainless-steel flow cell, all parts were soaked in 5% NaOCl (sodium hypochlorite) solution for 4 h, then rinsed with sterilized Millipore™ ultra-pure water and assembled under UV light. Then 70% ethanol in filtered sterile water was delivered to the flow cell for 5 min. All parts of the flow cell are assembled aseptically and placed into the system line.
Four glass tubes (3.5 mm diameter, 20 mm long) were also connected, in this system, both up-stream and down-stream of the flow cell. The surface area of the two glass tubes was approximately the same as the surface area of the cover glass in the flow cell. At each sample time, two glass tubes were removed, placed in 1 ml PBS solution, sonicated (20 kHz with a power output of 40 W) and the bacterial cell numbers per surface area were determined. Efficiency experiments showed that sonication of the glass tubes removed all of the adherent cells in one application. Destructive samples of the glass tubes were used to generate cell numbers/area in order to verify cell numbers per area determined by microscope non-invasively in the sealed flow cell.
Two strains (GFP- and RFP-containing strains) were cultivated individually in batch culture overnight. Pure suspended culture cells in exponential phase were centrifuged at 8,000 × g, washed, re-suspended, and centrifuged again. Then, cultures were diluted to known cell concentrations (107 cells/ml). Flow cells were then inoculated either with 107 cells/ml pure strain or with blended GFP and RFP strains in 1:1 or 1:3 proportions. The volumetric flow rate was set at 120 ml/h, and the cell suspensions were recycled (no effluent) through the flow cells for 2–3 h. After inoculation, LB or sterile modified FAB chemically defined media with 129 mg/l sodium citrate as the sole carbon source was supplied at a constant flow rate of 120 ml/h using a peristaltic pump. Biofilms that developed on the surface of the flow cells were examined periodically throughout an experiment. The flow cell was clamped at both ends and placed on the microscope stage for imaging.
Fluorescent microscopy and image analysis
Two different types of microscopes were used in this study: The first was an Axioskop2™ epi-fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a xenon bulb light source and two filter sets for FITC (ABS: 490–494 nm, emission: 517 nm) and Texas Red (ABS: 595 nm, emission 620 nm). An Optronics CCD camera (Optronics, Model S99802) controlled by MagnaFire™ (Optronics® California. USA) software was connected to the epi-fluorescence microscope, which generated two-dimensional digital color pictures. The second microscope was a Zeiss confocal laser scanning microscope LSM510 (Jena, Germany) mounted on an Axiovert 100 M inverted microscope. A Plan APO 100× oil objective with a numerical aperture of 1.4 or a C-APO 40× water immersion lens with a numerical aperture of 1.2 were used for obtaining cell images expressing green and red fluorescent proteins.
A “track” is a data-recording channel where excitation and emission wavelength information observed at specific values are stored. Two “tracks” were used to collect signals from the red and green proteins separately. Their composite superimposed images were constructed from each track image collected at different depths. “Track one” was set up at the excitation wavelength of 488 nm (argon laser) and a 505–530 nm band—pass (BP) emission filter to detect GFP-expressing bacteria. “Track two” was excited at 543 nm with an HeNe laser and a 590 long—pass (LP) emission filter to recognize DsRed-expressing bacteria. The spectra of GFP and DsRed have narrow emission bands with less crosstalk spectra, so the green color displayed by GFP and red color displayed by DsRed can be easily distinguished. A combination of GFP and RFP dual-color labeled studies within a single biofilm is possible with negligible overlap between emissions. All image analysis related to biofilm fluorescent density calibration was carried out using “Image J” version 1.30v software (Research Services Branch, National Institute of Mental Health, Bethesda, Maryland; http://rsb.info.nih.gov/ij/) and “Scion Image” based on NIH Image for Macintosh (The National Institutes of Health; http://www.scioncorp.com). To satisfy our goal of quantifying cell numbers from fluorescent densities, all of the control parameters in the image-analysis software were maintained constant between each picture.
“Density” is defined by the gray value within an image. The Image J software was used to calculate the area, mean density, integrated density, maximum and minimum grey value in each individual slice of the biofilm. In all experiments, images were acquired at several random axial (direction of fluid flow) positions at the upper, middle, and down stream portions of the flow channel. Biofilm in the areas near the inlet and outlet of the flow channel often displayed different topographical features due to hydrodynamic ‘entrance’ effects. At each axial position, image ‘stacks’ were taken at 1.0 to 2.0-μm-depth intervals through the biofilm; the number of images in each stack varied according to the thickness of the biofilm. A minimum area of at least 100,000 μm2 should be investigated in order to obtain representative data of P. fluorescens biofilms [18], so three (when applied 40× water immersion lens) to 12 (when applied 100× oil objective) image slices covering a total area of 1.0 × 105 μm were acquired for each flow chamber at different axial positions. A small Matlab computer program was used to calculate “integrated densities” for the large amounts of image data (not shown). All of the images obtained from epi-fluorescence microscopy and CLSM were digitally separated using the “x–y split” tool bar in Zeiss LSM 5 image browser into one channel for images of GFP or one channel for DsRed. The same method was used to calculate the number of cells in the biofilm for each strain.
All fluorescent densities and cell-number calibrations were carried out for each of the single slices with a sole signal (only GFP or only RFP). At early stages in biofilm development, integrated fluorescent densities for the entire image were measured along with cell numbers/area for each strain. With these values, “per cell” fluorescence was calculated for both GFP and RFP strains in every slice. These linear per-cell fluorescence correlations are used to predict cell-number concentrations from total image fluorescence collected from thick biofilms at latter stages of an experiment.
Results and discussion
Expression of GFP and DsRed in P. putida cells
GFP is a stable protein and persists under 60°C within a pH range of 2 to 12. Ideal as a reporter protein since GFP requires no substrate, its average maturation time is 45 min based on the different GFP mutants [25]. Conversely, DsRed (RFP) a mutant fluorescent protein of a different wavelength suffers with long maturation times and intra-cellular aggregation [22, 23]. Calibrating cell numbers determined from GFP observations against cell numbers determined from observation after staining the same cells with Live/Dead BacLight™ Bacterial Viability stain in liquid cultures indicated that only 90% of all cells exhibited GFP green fluorescence. Consequently, the maximum error in using expressed GFP alone to quantify cell numbers non-invasively would be about 10% (online SI figure S1).
Destructive and non-invasive measurement methods
During the early stages of an experiment, individual GFP-containing cells were counted directly from the images observed by epi-fluorescence microscopy. However, at some point in any experiment, images would contain too many adherent cells too close together to distinguish individual cells, thus obviating the direct counting of cells in the biofilm. Cell numbers in the biofilms were then determined non-invasively by collecting the total fluorescent density in an image, then dividing by the average fluorescent density of a single cell determined earlier in an experiment. This non-invasive approach was calibrated by directly measuring the cell areal concentration of biofilms samples destructively removed by sonication from glass tubes placed downstream from the flow cell system.
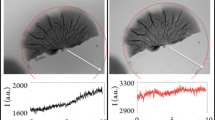
Total image fluorescent density was measured by three different methods: (1) “Mean density”, which is the sum of the gray values of all the pixels in the selection divided by the number of pixels, as provided directly by Scion Image; (2) “Integrated density”, provided by Image J or Scion Image software, is the sum of the gray value within a selection, minus the background; and (3) “Integrated density” is calculated from the “plot profile” minus the background sub-routine of Image J and Matlab software for density integration along with the X distance. Using all three methods, total densities determined by non-invasive fluorescent exhibited a linear relationship with total cell number #/area up to a certain limit (online SI figure S2). The mean bacterial fluorescent intensity of a single cell could be derived from the linear correlation between direct cell number counts and the total intensity. The calculation approach was correlated by directly measuring the areal concentration of cells in biofilm samples destructively removed from flow cell surfaces. Two glass tubes, at the influent or effluent ends of the flow cell system, simulated environmental conditions similar to those at the glass surface in the flow cell. In addition, these two glass tubes were built with the same total area as the coverslip in the flow cell.
Figure 2 compares the adherent cell concentrations (cell #/area) derived from the four different methods. Only method 3 results compared favorably to cell concentrations determined directly from destructive samples biofilm for all 3 days. As a result, method 3, the “integrated density” calculated from “plot profile” with the background subtracted was chosen for the non-invasive measurement of the adherent cell concentrations.
Comparison of adherent cell concentrations (cell #/cm2) derived by four different methods. The original X–Y images of Ppu2440 (miniTn7PA1/03/04egfp-a) biofilm were taken with an epifluorescent microscope equipped with a ×100 oil immersion lens. Methods 1–3 are image-analysis methods that employ subtle, different ways of processing the original adherent cell image. Biofilm cell concentrations were also determined by destructive removal of biofilm and assessment of the suspended cell numbers
Method 1 did not take the image background into consideration. However, each image slice has its own fluorescent background, so this method overestimates total cell densities and numbers. Method 2 showed appropriate comparison with destructive biofilm samples on day 1 and day 2. However, a large deviation with the destructive biofilm samples was observed on day 3 data. The values of the image background were the main reason for this discrepancy. Method 2 measures the background and counts the integrated density using the Scion Image software directly. When the biofilm was well developed and cell number concentration was large, the entire image area is covered with fluorescent cells. The main brightness on each of the images is mistakenly considered the background. In contrast, the bright images with large background values were inverted in method 3 to obtain reasonable background values. Adherent cell numbers counted by method 3 showed comparable results to destructive biofilm samples.
Quantifying mixed-strain biofilm development
Figure 3 presents correlations for total image fluorescence versus cell number areal concentration for the mixed-strain biofilms of GFP and RFP, respectively, under the 100× oil immersion lens. Correlation R 2 values are greater than 0.98 for both the GFP and RFP strains. Similar correlations with R 2 > 0.98 were also obtained with a 40× objective (data not shown).
Integrated densities as a function of cell number per area. Linear trendlines were added to fit the sample data. Fitting equations and R-squared values were displayed for each of the fitting equations. a GFP signals, b RFP signals. Per area = 90 × 90 μm. The images of Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) mixed-strain biofilms were taken with a CLSM equipped with a ×100 oil immersion lens. Red and green signals were split and analyzed to the above linear fitting lines, respectively
Biofilm development was investigated using GFP- and RFP-containing mixed strains. Spatial relationships between Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) strains in a developing mixed-strain biofilm are shown in the micrographs of Fig. 4 taken with a CLSM with a 40× magnifying water immersion lens. This experiment was inoculated with a 1:1 ratio of the two strains. As shown in Fig. 4, RFP and GFP containing mixed biofilms formed distinct structures as time progressed. On days 1, 2, and 4, most of the cells were seen only as loose discrete structures. These images show that separate green or red fluorescent microcolonies were formed initially from individual red and green fluorescent cells. Results indicate that the initial individual microcolonies that formed continued to grow as separate pure strain clusters instead of a uniform mixing of both strains in a cell aggregation process. On days 8, 10, and 12, the mixed strains continued to form biofilm clusters that merged together to form the biofilm. While the majority of fluorescent emission overlap between GFP and DsRed was eliminated by the appropriate setting of the emission filters, a small amount of fluorescent overlap is recorded as yellow color (shown in Fig. 4d, e, and f).
X–Y images of Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) mixed strains in a developing biofilm studied by the use of CLSM equipped with a C-APO ×40 water immersion lens. Images a, b, c, d, e, and f were collected on day 1, 2, 4, 8, 10, and 12 days of continuous biofilm formation, respectively. These (X, Y) images were taken at an axial position (X) in the flow cell about 1.9 cm from the inlet, which is almost at the center of the flow cell. (X, Y) Images b, c, d, e and f were the brightest images chosen from all of the (Z) slices in each stack. The total thickness of the stacks for image b, c, d, e, and f were 10, 24, 40, 65, and 109 μm, respectively. Images of b, c, d, e, and f were located at a depth of around 8.0 μm in the Z direction. Image A did not count in the densities comparison. The depth of this slice was not measured since the biofilm was started only for 1 day and the depth of the biofilm was smaller than 5 μm
A second flow cell experiment was carried out with a starting inoculation of Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) bacteria cells of 3:1 in PBS solution. Figure 5 displays an entire Z stack of X–Y horizontal sections (presented in the “Gallery Mode”) taken on day 10 of the experiment. The majority of Ppu2440 (miniTn7Pmtdsred) (red) cells concentrated near the glass substratum. The stratification of strains within the biofilm may be attributed to subtle differences in growth rate, adhesion abilities, and local differences in substratum surface chemistry [10]. Despite the higher starting inoculation concentration of the DsRed strain, cell numbers of the GFP strain decreased only slightly throughout the biofilm, while cell numbers of the RFP strain decreased dramatically. The GFP strain dominates all X–Y images at altitudes ≥30 μm above the substratum (Fig. 6). As biofilms accumulate and extend away from the substratum, cells at the bottom of a film have had a longer period in “residence” than those that are newly produced at the progressing edge of the biofilm. Since our DsRed reporter protein requires at least 24 h to mature [19], there will be a biased gradient of DsRed strains at the bottom relative to DsRed cells in the upper regions of the biofilm. Ideally, to correct this bias would require two fluorescent markers that mature at the same rate.
Display windows of a Z stack in X–Y horizontal sections of a Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) mixed-strain biofilm taken with a CLSM equipped with a C-APO ×40 water immersion lens. The whole 3-D stack was collected on day 10 of continuous biofilm formation. All of the image slices showed in this stack were taken at an axial position in the flow cell about 1.9 cm from the inlet in the center of the flow cell. Each slice represents images at different marked depth above the cover glass of the biofilm. Scale bar 20 μm
Areal cell concentration in biofilm distributed with the different thickness on day 10 of a Ppu2440 (miniTn7Pmtdsred) and Ppu2440 (miniTn7PA1/03/04egfp-a) mixed-strain biofilm taken with a CLSM equipped with a C-APO ×40 water immersion lens. Cell concentration was quantified for GFP strain and RFP strain, respectively. The CLSM images of this stack were collected on day 10 of continuous biofilm formation. The image stack was taken at an axial position in the flow cell about 1.9 cm from the inlet in the center of the flow cell. All of the slices in this stack were scanned automatically by CLSM along with the different thickness of the biofilm
Conclusions
Our goal here was to develop a non-invasive microscope quantification technique to determine biofilm cell concentrations in pure and mixed-population biofilms. The method was calibrated with two green fluorescent protein (GFP) mutants in a continuous flow cell system.
Epi-fluorescence and confocal laser scanning microscopes were used to detect the total image cell fluorescent intensities and adherent cell numbers/area at early stages of biofilm development when individual cells were still discernable. From these two measures, a per-cell fluorescence value was determined. Local and overall cell bacterial concentrations/area within a biofilm were then quantified at latter stages of development, based on this per-cell fluorescence value. Estimates of late-stage biofilm cell concentrations determined with this non-invasive technique were correlated with direct cell counts on destructive samples.
The technique of fluorescent correlations to cell numbers determined in a pure-strain system were used directly without change in mixed-strain cultures since these mixed-biofilm images can be separated and exported as pure-strain images. CLSM is naturally more reliable for quantification of thick biofilms than epi-fluorescent microscopy. Use of this method to estimate local dynamic plasmid segregational loss and plasmid conjugation transfer kinetics will be reported in a subsequent manuscript.
References
Andersen JB, Sternberg C, Kongsbak-Poulsen L, Petersen-Bjorn S, Givskov M, Molin S (1998) New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246
Beaudoin DL, Bryers JD, Cunningham AB, Peretti SW (1998) Mobilization of broad host range plasmid from Pseudomonas putida to established biofilm of Bacillus azotoformans. I: experiments. Biotechnol Bioeng 57:272–279
Banks MK, Bryers JD (1991) Bacterial species dominance within a binary culture biofilm. Appl Environ Microbiol 57:1974–1979
Brading M, Jass J, Lappin-Scott HM (1995) Dynamics of bacterial biofilm formation. In: Lappin-Scott HM, Costerton JW (eds) Plant and microbial biotechnology research series, no. 5. Microbial Biofilms. Cambridge University Press, Cambridge, pp 46–63
Bryers JD (ed) (2000) Biofilms-II. Wiley-Liss Publishers, New York
Bryers JD (1988) Modeling biofilm accumulation. In: Bazin MJ, Prosser JI (eds) Physiological models in microbiology, vol II. CRC Press, Boca Raton, pp 109–144
Chalfie M, Tu Y, Euslichen G, Ward W, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263:802–805
Characklis WG, Marshall KC (eds) (1990) Biofilms. Wiley, New York
Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38
Cowan SE, Gilbert E, Khlebnikov A, Keasling JD (2000) Dual labeling with green fluorescent proteins for confocal microscopy. Appl Environ Microbiol 66:413–418
Crameri A, Whitehorn EA, Tate E, Stemmer WPC (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14:315–319
Eberl L, Schulze R, Ammendola A, Geisenberger O, Erhart R, Sternberg C, Molin S, Amann R (1997) Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol Lett 149:77–83
Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, Huang C-T, Fredericks J, Burnett S, Stewart PS, McFeters G, Passador L, Iglewski BH (1999) Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol 34:1082–1093
Huang C-T, Peretti SW, Bryers JD (1993) Plasmid retention and gene expression in suspended and biofilm cultures of recombinant E. coli DH5α (pMJR1750). Biotechnol Bioeng 41:211–220
Huang C-T, Peretti SW, Bryers JD (1994) Effects of medium carbon-to-nitrogen ratio on biofilm formation and plasmid stability. Biotechnol Bioeng 44:329–336
Huang C-T (1993) Plasmid retention and gene expression in bacterial biofilm cultures, PhD dissertation, Duke University, Durham, NC
Heydorn A, Ersboll BK, Hentzer M, Parsek MR, Givskov M, Molin S (2000) Experimental reproducibility in flow-chamber biofilms. Microbiology 146(Pt 10):2409–2415
Korber DR, Lawrence JR, Hendry MJ, Caldwell DE (1993) Analysis of spatial variability within Mot+ and Mot− Pseudomonas fluorescens biofilms using representative elements. Biofouling 7:339–358
Lambertsen L, Sternberg C, Molin S (2004) Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol 6(7):726–732
Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE (1991) Optical sectioning of microbial biofilms. J Bacteriol 173:6558–6567
Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101–109
Maksimow M, Hakkila K, Karp M, Virta M (2002) Simultaneous detection of bacteria expressing GFP and DsRed genes with a flow cytometer. Cytometry 47:243–247
Nancharaiah YV, Wattiau P, Wuertz S, Bathe S, Mohan SV, Wilderer PA, Hausner M (2003) Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl Environ Microbiol 69(8):4846–4852
Silverstone LM, Johnson NW, Hardie JM, Williams RAD (1981) The formation, structure and microbial composition of dental plaque. In: Silverstone LM, Johnson NW, Hardie JM, Williams RAD (eds) Dental caries: aetiology, pathology and prevention. Macmillan Press, London, pp 70–102
Skillman LC, Sutherland IW, Jones MV, Goulsbra A (1998) Green fluorescent protein as a novel species-specific marker in enteric dual-species biofilms. Microbiology 144(Pt 8):2095–2101
Socransky SS, Haffajee AD (1992) The bacterial etiology of destructive periodontal disease: current concepts. J Periodontal 63:322–331
Stewart PS (1994) Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38:1052–1058
Acknowledgments
Microscopy was carried out at the Center for Biomedical Imaging, University of Connecticut Health Center, with the assistance of Susan Kreuger. This work was support by the NSF Grant BES-0121203.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, H., Bryers, J.D. Non-invasive method to quantify local bacterial concentrations in a mixed culture biofilm. J Ind Microbiol Biotechnol 37, 1081–1089 (2010). https://doi.org/10.1007/s10295-010-0756-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0756-z