Abstract
Moenomycin, a natural phosphoglycolipid product that has a long history of use in animal nutrition, is currently considered an attractive starting point for the development of novel antibiotics. We recently reconstituted the biosynthesis of this natural product in a heterologous host, Streptomyces lividans TK24, but production levels were too low to be useful. We have examined several other streptomycetes strains as hosts and have also explored the overexpression of two pleiotropic regulatory genes, afsS and relA, on moenomycin production. A moenomycin-resistant derivative of S. albus J1074 was found to give the highest titers of moenomycin, and production was improved by overexpressing relA. Partial duplication of the moe cluster 1 in S. ghanaensis also increased average moenomycin production. The results reported here suggest that rational manipulation of global regulators combined with increased moe gene dosage could be a useful technique for improvement of moenomycin biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Significant effort has been put into understanding the biology of secondary metabolism of actinomycetes in order to accelerate and simplify the generation of industrial overproducers from typically low-producing wild-type strains [3, 5]. The regulation of antibiotic production is a complex multigene trait where genetic circuits of different rank, and environmental and intracellular cues are intricately combined to tune the expression of certain metabolic pathways [4, 13]. Although the general architecture of such regulation seems to be conserved across species, its effects on production of a given molecule cannot always be predicted and have to be addressed experimentally [7].
We are interested in moenomycin A (MmA; Fig. 1), a phosphoglycolipid antibiotic that inhibits peptidoglycan biosynthesis. For almost four decades MmA has been used as an animal growth promoter without a notable rise in MmA-resistant microflora. Now, when chemotherapy of human infections faces a serious shortage of efficient drugs, MmA has attracted the attention of both chemists and biologists as a promising model for the development of novel classes of antibiotics to treat multidrug-resistant pathogens [2, 17, 31, 33]. Exploiting its promise requires having access to significant quantities of moenomycin intermediates for further modification. We have previously identified the genes for MmA biosynthesis in the producing organism, S. ghanaensis, and have shown that a variety of Mms can be successfully produced in S. lividans TK24 [23]. However, low Mm production levels in both the natural producer, S. ghanaensis, and the heterologous host call for the development of better Mm-producing strains. Since no apparent regulatory genes were found in the MmA biosynthetic gene (moe) cluster [21], and the genetic factors that control its production are not understood, we have examined several different strategies to improve biosynthesis in model moenomycin-producing strains. Our initial studies, reported below, support the idea that manipulating global antibiotic regulatory genes and gene dosage in suitable strain backgrounds can lead to useful improvements in production levels for other studies.
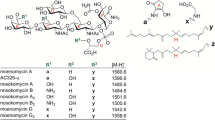
Structures of moenomycins relevant to this study. MmA and compound 1 are accumulated by S. ghanaensis strains, while the production of compound 2 is directed by cosmids moeno38-5 and moeno38-1 in heterologous hosts [21]
Levels of moenomycin production by various streptomycete strains. a B. cereus growth inhibition around paper discs saturated with methanol extracts from 100 mg (wet weight) of biomasses of different Mm producers and control strains: S. albus J1074 (1), S. coelicolor M145 (2), 2 μg of MmA (3), M512 moeno38-5+ (4), M145 moeno38-5+ (5), J1074 moeno38-5+ (6), R1 moeno38-5+ (7), R1 moeno38-5+ afsS + (8), R1 moeno38-5+ relA + (9), TK24 (10), TK24 moeno38-5+ (11), 1326 moeno38-5+ (12), S. ghanaensis (13), S. ghanaensis relA + (14). b Moenomycin productivity of heterologous strains expressing cosmid moeno38-5. Column labels: TK24, 1326—rpsL and wild-type strains of S. lividans, respectively; M145, M512—wild-type and Act−Red− strains of S. coelicolor; J1074, R1—parent and MmA-resistant strains of S. abus, respectively; R1afsS and R1relA—R1 strains overexpressing afsS and relA genes, respectively. c Moenomycin productivity of different S. ghanaensis strains. Column labels: ghana—wild-type (ATCC14672); ghana afsS, ghana relA, and ghana 38-5—S. ghanaensis strains overexpressing afsS, relA, and cosmid moeno38-5, respectively. The Mm production values of relA-expressing strains and S. ghanaensis carrying duplicated moe cluster 1 are statistically significant, since they did not overlap with 2σ range of the control strains
Materials and methods
Bacterial strains and plasmids
Escherichia coli DH5α (Life Technologies, Carlsbad, CA, USA) was used for routine subcloning. E. coli ET12567 (pUB307) was used to perform intergeneric conjugation from E. coli to Streptomyces strains. Bacillus cereus ATCC19637 was used as a test-culture for antibiotic disc diffusion assays. Commonly used S. coelicolor M145, M512, S. lividans TK24, 1326 [14], and S. albus J1074 [6] strains were tested here as Mm producers. Plasmids pKD46 and pKD4 were a gift from Prof. J. Beckwith (Harvard Medical School, USA); they were used to carry out RedET-mediated gene replacement as described in following paragraph. Streptomyces expression vectors pKC1218E, pIJ6902, pSOK101 and cosmid moeno38-1 were described previously [13, 21]. S. coelicolorrelA expression construct pIJ6085was provided by Prof. M. Bibb (JIC, Norwich, UK). List of recombinant molecules constructed and used along with resulting strains are listed in Table 1.
Culture conditions and general methods
Standard genetic techniques for E. coli and Streptomyces and for DNA manipulations were used as described [14, 25]. Streptomyces strains were grown in liquid R5A [26] or TSB media [14] for Mm production. E. coli and Bacillus strains were grown in LB supplemented with appropriate antibiotics. Oatmeal medium [18] was used to obtain spores of streptomycetes and to plate intergeneric matings. Where needed, streptomycete strains were incubated in the presence of antibiotics: kanamycin (50 μg ml−1), apramycin (50 μg ml−1), or hygromycin (100 μg ml−1). T4 DNA ligase and restriction enzymes were used according to the recommendations of suppliers (NEB, Beverly, MA; MBI Fermentas, Burlington, ON, Canada). Other DNA manipulations were carried out following standard procedures specified by the manufacturers (NEB; MBI Fermentas; Invitrogen, Carlsbad, CA, USA; Bio-Rad, Hercules, CA, USA).
Moenomycin extraction, purification, and quantitative analysis
One-hundred-milliliter flasks with 15 ml of seed medium (TSB) were inoculated with frozen spore suspensions (2 × 105 cfu). The flasks were incubated at 28°C for 48 h on a rotary shaker at 180 rpm. One ml of the resulting seed broth was used to inoculate the 300-ml flasks with 30 ml of fermentation medium (R5A for heterologous hosts and TSB for S. ghanaensis strains). The fermentation was carried out on a rotary shaker at 180 rpm for 5 days at 30°C. The fermented broth was centrifuged at 3,500 rpm for 10 min. The mycelium was washed twice with distilled water. Mm was extracted by stirring the biomass (1 g, wet weight) with 3 ml of methanol for 12 h. The extract was concentrated in vacuo and diluted to the final volume of 300 μl. Then it was either directly used for antibiotic disc diffusion assay or purified for LC–MS analysis according to the described procedure [21–23]. For the antibiotic diffusion assay, paper discs (∅5 mm, Whatman, Maidstone, UK) were impregnated with a portion of the extract and dried at 37°C for 1 h. Discs were placed on the plates with 0.7% soft agar containing B. cereus (108 cfu). The plates were incubated at 4°C for 1 h and then at 37°C for 17 h. Relative changes in moenomycin production by various strains were determined with the help of LC–MS as described [23]. For heterologous strains, the quantity of compound 2 (Fig. 1) was determined, whereas for S. ghanaensis strains Mm productivity is the sum of production of MmA and its intermediate 1 (Fig. 1). Mean values were found from at least four biological repeats and standard deviations (σ) are shown in Figs. 2 and 3. Several control experiments were carried out to ensure that multistep purification and analysis process did not distort the measurements. Samples that contain increasing concentrations of MmA (in the 0.003–10 μM range) were run through LC–MS and shown to give proportional increases in MmA-specific mass-peak area. Pure MmA was used as an internal standard for Mm extracts from heterologous hosts. The only samples that showed essentially identical MmA: 2 ratio upon LC–MS analysis were then taken into account. Under our LC–MS conditions [23], 1 AU of moenomycin productivity corresponds to 1580.6-Da peak area resulted from injection of 100 pM MmA; 105 AU corresponds to 10 μM MmA.
Time course for biomass and Mm accumulation by S. albus R1 38-5 strains with and without relA. Solid lines biomass production, dashed lines specific Mm productivity, open diamonds R1 38-5 strain, solid rectangles R1 38-5 + relA strain. The productivity values are statistically significant as mentioned in the legend of Fig. 3
Verification of the strains containing cosmid moeno38-5
Kanamycin- and hygromycin-resistant transconjugants carrying cosmid moeno38-5 were checked for the stability of inheritance of antibiotic resistance phenotype. Two hundred clones of each transconjugant were checked after three passages under nonselective conditions and all were found to retain the expected phenotype. PCR was employed to prove the presence of moeno38-5 in the chromosomes of streptomycete hosts. For this purpose, we used primers moeO5up and moeO5rev (Table 1S, Electronic Supplemental Materials, ESM) specific for moeO5 gene and those used to amplify attR ϕC31 [28], the “right” arm of moeno38-5 integration. The amplicons of expected size (1.3 and 0.6 kb for moeO5 and attR ϕC31, respectively) were amplified from all transconjugants in case of both primer pairs.
Plasmid construction
For afsS overexpression
pIJ4112 [11] was treated with PstI and the resulting 2.9-kb fragment containing the entire afsS and a part of afsR genes was cloned into PstI-digested vector pKC1218E [21]. The final construct (pKCafsS) contained gene afsS under the control of ermEp*.
For moeO5p-neo fusion
A 1.5-kb DNA fragment carrying the fd terminator, MCS, and promoterless neomycin resistance gene neo was amplified from pMO9 using primers terforNheI and neorevMfeI [24], and cloned into pGEM T-easy vector (Promega, Madison, WI, USA) to give pGN1. The 515-bp DNA fragment adjacent to the moeO5 gene that includes the first 35 bp of the ORF was amplified with primers O5PFHindIII and O5PRXbaI (Table 1S, ESM) from cosmid moeno38-1 [21] and cloned into pSTBlue1 Acceptor vector (Novagen, Whitehouse Station, NJ, USA). The promoter region was retrieved with HindIII-XbaI and cloned into respective sites of pGN1. Obtained plasmid pGNO5P was digested with NheI and MfeI and the neo cassette fused to moeO5p was cloned into EcoRI/XbaI sites of pOOB5 [21] to give pONO5P. All constructs mentioned in this study were verified by sequencing at the Biopolymer Facility (Harvard Medical School).
For moeO5p titration experiments
A 0.5-kb DNA fragment upstream of the moeO5 start codon was amplified with aforementioned primers O5PFHindIII and O5PRXbaI. The resulting amplicon was digested with HindIII and XbaI and cloned into respective sites of high-copy number pIJ101-based vector [14] pSOK101 to give pSOKmoeO5p.
Cosmid moeno38-5
Cosmid moeno38-5 was constructed to ensure that the moe cluster-flanking 18-kb segment of S. ghanaensis chromosome (Fig. 4) does not cause any possible effects on Mm production by heterologous strains. The kanamycin resistance gene from plasmid pKD4 [8] was amplified with primers 38start-KD4 and moeA5-P3 (Table 1S, ESM). The resulting amplicon was used to replace the moeA4moeB5 gene pair as well as the entire 18-kb “left arm” of moeno38-1 (Fig. 4) using the described λRed approach [8]. We did not evict the kanR gene region from moeno38-5 because it did not exert any negative effects on MmA production. The replacement of the moeA5 and moeB5 genes with kanR in moeno38-5 was confirmed via diagnostic PCR (primers P2-KD4 and alsrev1 (Table 1S, ESM). Levels of Mm production by moeno38-5+ and moeno38-1+ S. lividans TK24 transconjugants were identical (data not shown), implying that no regulatory or structural moe genes were present in the deleted 18-kb region.
Semiquantitative RT-PCR
Total RNA samples were isolated after 4 days of S. albus growth in R5A according to instructions of the manufacturer of the RNeasy kit (Qiagen, Valencia, CA). To avoid DNA contamination, samples were treated with RQ1 DNase (Promega). RNA concentration and purity were determined by measuring the ratio of OD260/OD280, and an equal amount of RNA from each studied strain was used for RT reactions. cDNA was obtained using AMV Reverse Transcriptase (Promega) and random hexanucleotide primers. PCR was performed using Go-Taq DNA polymerase (Promega) and primer pairs specific to moeO5 and moeE5 genes (350-bp segments, Table 1S, ESM). As a positive control, the rrnA primer pair specific to 16S rRNA of Streptomyces coelicolor (380-bp segment) was used. Negative controls were carried out with rrnA primers to confirm the absence of contaminating DNA in the RNA preparations. PCR products were analyzed by electrophoresis in 1.2% agarose gels, and band intensity was established using ImageJ1.36b software (National Institutes of Health, Bethesda, MD, USA).
Screening for global regulators in genomes of S. ghanaensis ATCC 14672 and S. albus J1074
To assess the presence of pleiotropic regulators of antibiotic production in the genomes of S. ghanaensis ATCC 14672 and S. albus J1074, draft genomic assemblies of these strains were downloaded from the Web site of Broad Institute (http://www.broad.mit.edu/annotation/genome/streptomyces_group/). The Open Read Frame (ORF) prediction was performed by a self-training application Glimmer3 [9] with default settings. Protein sequences of 20 known global regulators of S. coelicolor A3(2) secondary metabolism were collected from GenBank with subsequent BLAST [1] comparison to the genomes of interest. The list of regulators and other details are given in the ESM. Apart from turning off “low-complexity filtering” in the BLAST suite, we have used default values. In a few cases, manual adjustment of predicted ORFs was needed (i.e., if the sequence alignment extended beyond ORF boundaries with high probability). The identity value in the table is derived from best high-scoring pair (HSP) from the BLAST output, not the whole proteins, however in most cases these coincide (or almost coincide). Due to the unfinished status of sequencing projects of both genomes, no data on exact location in the chromosome or fixed locus tags for the predicted regulators can be presented.
Results
Generation and analysis of novel heterologous hosts for Mm production
Although novel MmA analogs were successfully isolated from recombinant S. lividans strains [21, 23, 33], their low productivity (up to 500 μg l−1) is a formidable challenge for purification of large amounts of Mms needed for various studies. Better heterologous moenomycin producers would significantly facilitate the chemoenzymatic synthesis of novel phosphoglycolipids. Since the generation of (and especially analysis of) Mm-producing strains is laborious, a simple method to find the optimal host(s) would be desirable. We reasoned that the efficiency of expression of a key gene for MmA production, moeO5 [23] in different strains could be used to sort them according to their ability to produce Mm. The activity of the moeO5 promoter has been established in promoter titration experiments. Particularly, the S. ghanaensis pSOKmoeO5p+ strain, carrying multiple moeO5p copies in trans, accumulated significantly smaller amounts of moenomycins in the mycelium [(0.3 ± 0.1) × 105 AU mg−1 dry biomass] as compared to the control strain [(1.4 ± 0.2) × 105 AU mg−1 dry biomass]. Hence, as-yet-unknown transcriptional activator(s) appear to interact with moeO5p and their activity is effectively “diluted” by plasmid-borne moeO5p. This result encouraged us to construct pONO5P that carries a transcriptional fusion of moeO5p to the promoterless kanamycin resistance gene neo. As a test case, we compared the kanamycin resistance levels that pONO5P confers to three strains widely used as heterologous hosts, namely S. coelicolor M512 (ΔactII-ORF4, ΔredD), S. lividans TK24, and S. albus J1074. The 10-μl aliquots containing approx. 2 × 104 cfu of three independent pONO5P+ transconjugants of each aforementioned strain were applied to TSB agar plates supplemented with different amounts of kanamycin (1, 2.5, 5, 7.5, 10, 15, 25, and 50 μg ml−1). Only S. albus grew at the highest kanamycin concentration tested (50 μg ml−1). Good growth of S. lividans TK24 was observed on TSB with 25 μg ml−1 of kanamycin, whereas S. coelicolor M512 grew on 2.5 μg ml−1. No growth of the M512 and TK24 control strains carrying empty vector, was observed at the lowest kanamycin concentration tested. In the case of the control J1074 strain, a slight growth was observed at 2.5 μg ml−1 of kanamycin. Next, cosmid moeno38-5 [carrying no additional segments of the S. ghanaensis genome except for the moeD5-moeGT3 portion of the moe cluster 1 (Fig. 4)], was introduced into different strains of S. albus, S. coelicolor and S. lividans J1074 to compare their Mm production levels. S. albus gave the highest Mm titer, while M512 had the lowest one. There was approximately a 30-fold difference in production levels between J1074 and M512, showing considerable variation between heterologous hosts (Fig. 2). Expression of the moeO5p-neo construct and Mm production in the strains being tested seem to correlate positively, therefore such reporter systems can potentially be used as a tool to screen for optimal strains and conditions for Mm production.
S. coelicolor is naturally resistant to MmA, as are most streptomycetes strains we have examined, but S. albus 1074 is sensitive, with a survival rate of 0.001% at 1 μg ml−1 of MmA (data not shown). Since MmA sensitivity could affect production levels in S. albus, we introduced cosmid 38-5 into the spontaneous MmA-resistant S. albus strain R1 (97% survival at 1 μg ml−1 of MmA; data not shown) and compared growth and Mm accumulation in the R1 and J1074 transconjugants. The resistant strain showed a negligible increase in Mm production. Since MmA is primarily accumulated inside of the cells [23] and its target is located on cell surface, it is conceivable that certain (low) levels of MmA production can be achieved even by streptomycetes very sensitive to exogenously added MmA.
Since cross-regulation of antibiotic production has been previously reported for S. coelicolor, we examined whether the heterologous production of Mms could be affected by the production of actinorhodin (Act) and undecylprodigiosin (Red). The Mm production levels in moeno38-5+ S. coelicolor M145 (Act+ Red+) were still low, but were reproducibly higher (three times) than in the Act- and Red-deficient variant moeno38-5+ M512 (Fig. 2a). Hence, some of the act/red genes or Act/Red themselves have a positive, albeit modest, influence on MmA biosynthesis.
We also investigated the influence of a mutation in the rpsL gene in S. lividans on Mm production levels. S. lividans TK24, used as the original heterologous host for Mm production, contains a point mutation (K43R) in the rpsL gene encoding ribosomal protein S12. This mutation is reported to enhance antibiotic production in various actinomycetes [27]. To determine the effect of this mutation on Mm production, we compared production of Mms in S. lividans TK24 (rpsL mutant) and the parent strain S. lividans 1326. Both antibiotic disc diffusion and LC–MS analyses (Fig. 2) showed that 1326 and TK24 accumulated comparable amounts of moenomycins. Hence, the rpsL K43R mutation does not significantly affect the level of Mm production.
In silico analysis and initial in vivo experiments show that certain global regulators may influence Mm production
The absence of pathway-specific regulatory genes associated with the moe cluster and its differential expression in various streptomycetes point to the possibility that some global antibiotic biosynthesis regulators govern MmA production. Such regulators are well studied in S. coelicolor [19] and we searched for their counterparts in partially sequenced genomes of S. ghanaensis and S. albus J1074. Our in silico analysis showed that both strains possess homologues of transcriptional factors, components of the A-factor signaling cascade and other proteins orchestrating secondary metabolism of S. coelicolor (Table 2S, ESM). Two S. coelicolor genes, afsS and relA, were chosen as a test case to obtain experimental evidence for their possible role in regulation of MmA production. These genes are known to cause the overproduction of secondary metabolites in S. coelicolor, S. lividans, and S. avermitilis [4, 11, 15, 30]. The exact function of afsS gene is unknown [16], while relA encodes (p)ppGpp synthetase [29]. Strain S. albus R1 moeno38-5+ relA + showed a twofold increase in Mm production level compared to S. albus R1 moeno38-5+ carrying empty vector (Fig. 2). Overexpression of the afsS gene in S. albus R1 moeno38-5+ increased Mm production negligibly (1.25-fold; Fig. 2a, b). We introduced the regulatory genes described above into S. ghanaensis ATCC14672 and studied their effects on MmA production. Gene afsS had no impact on Mm biosynthesis, while the relA + strain produced roughly twofold more Mm than control strain S. ghanaensis (pIJ6902) (Fig. 2). Of all combinations tested, relA expression in S. albus had the biggest positive impact on Mm production, and we investigated this case in more detail. Although growth curves of relA + and control strains were virtually identical, there is marked increase in specific Mm productivity of relA + variant starting from the end of the 4th day, and declining slightly on the 6th day. This is paralleled by increased transcription of two key moe genes, moeO5, and moeE5, on the 4th day of fermentation of the 38-5+ relA + strain, as judged by semiquantitative RT-PCR (Fig. 5).
RT-PCR analysis of moeO5 and moeE5 transcription in S. albus R1 38-5+ and R1 38-5+ relA + strains during Mm production phase (96 h). Lanes: 2, 5—rrnA transcript from parent and relA + strain, respectively; 3, 4—moeE5 and moeO5, respectively, from R1 38-5+ relA +; 6, 7—moeE5 and moeO5, respectively, from R1 38-5+; 8—negative control (rrnA amplification from RNA sample in absence of RT); 1—1-kb ladder
Increased moe gene dosage influences MmA production by S. ghanaensis
Massive amplification of antibiotic biosynthesis genes is often one of the outcomes of empirical strain improvement programs [10, 32]. To test whether an increased dosage of moe genes could be used as a tool to enhance MmA biosynthesis, we integrated cosmid moeno38-5 into the S. ghanaensis attB ϕC31 site. The resulting strain thus carries at least two copies of most of moe cluster 1 genes. We revealed that introduction of moeno38-5 into S. ghanaensis led, on average, to a twofold increase in Mm production (Fig. 2c).
Discussion
MmA is considered a promising lead to combat vancomycin- and methicillin-resistant pathogens because of its high potency, uniqueness of structure, and mode of action [12, 17, 20]. Better knowledge of the factors that limit Mm production by streptomycetes would certainly help resolve the issue of intrinsically low Mm yields. Here we describe a simple reporter system that may be used to find an efficient heterologous host for the production of moenomycins. In a test case, screening of several well-studied streptomycetes turned up heterologous host, S. albus, as being better than S. lividans or S. coelicolor in terms of Mm production level and ease of antibiotic purification (because of the fast and dispersed growth and absence of pigmented metabolites). Of note, we revealed a dramatic difference in Mm-producing capacities of several species being tested, highlighting the importance of host-specific genetic factors for antibiotic production. More extensive screening with the help of the described reporter system may reveal even better strains.
We also addressed for the first time the possibility of the use of heterologous regulatory elements for MmA improvement. As the relA expression shows, at least some of the well-known pleiotropic regulatory genes might exert significant effects on Mm production, probably through direct or indirect modulation of expression of structural moe genes. This result encourages further investigations on the possible use of S. coelicolor global regulators to enhance Mm biosynthesis. Finally, duplication of moe cluster segments may increase antibiotic yields. We envision that proper combination of the discovered genetic factors with optimized fermentation media and purification procedures can significantly improve the production of novel moenomycins by recombinant strains. It is not known at the moment whether positive effects observed in our work (e.g., relA expression and moe gene dosage) will be additive. Although the results presented here are for model/wild-type strains, their application to known industrial strains or other potential heterologous hosts is anticipated to exert similar effects on Mm production.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baizman ER, Branstrom AA, Longley CB, Allanson N, Sofia MJ, Gange D, Goldman RC (2000) Antibacterial activity of synthetic analogues based on the disaccharide structure of moenomycin, an inhibitor of bacterial transglycosylase. Microbiology 146:3129–3140
Baltz RH (1998) Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol 6:76–83
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Champness W (2000) Actinomycete development, antibiotic production and phylogeny: questions and challenges. In: Brun YV, Skimkets LJ (eds) Prokaryotic development. Am Soc Microbiol, Washington, DC, pp 11–31
Chater KF, Wilde LC (1980) Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol 116:323–334
Chen L, Lu Y, Chen J, Zhang W, Shu D, Qin Z, Yang S, Jiang W (2008) Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl Microbiol Biotechnol 80:277–286
Datsenko K, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Fedorenko V, Golets L, Yu Demydchuk, Kriugel H (1998) Analysis of genome rearrangements in Streptomyces kanamyceticus mutants. Antibiot Khimitoter (Rus) 43:14–19
Floriano B, Bibb M (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21:385–396
Goldman RC, Gange D (2000) Inhibition of transglycosylation involved in bacterial peptidoglycan synthesis. Curr Med Chem 7:801–820
Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN (2005) Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol 58:1276–1287
Kieser T, Bibb M, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Lee J, Hwang Y, Kim S, Kim E, Choi C (2000) Effect of a global regulatory gene, afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng 89:606–608
Lian W, Jayapal KP, Charaniya S, Mehra S, Glod F, Kyung YS, Sherman DH, Hu WS (2008) Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2). BMC Genomics 9:56
Lovering AL, de Castro LH, Lim D, Strynadka NC (2007) Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405
Luzhetskyy AM, Ostash BO, Fedorenko VO (2001) Intergeneric conjugation Escherichia coli–Streptomyces globisporus 1912 with using of integrative plasmid pSET152 and its derivative. Rus J Genet 37:1340–1347
McKenzie NL, Nodwell JR (2007) Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol 189:5284–5292
Ostash B, Walker S (2005) Bacterial transglycosylase inhibitors. Curr Opin Chem Biol 9:459–466
Ostash B, Saghatelian A, Walker S (2007) A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol 14:257–267
Ostash B, Makitrinskyy R, Walker S, Fedorenko V (2009) Identification and characterization of Streptomyces ghanaensis ATCC14672 integration sites for three actinophage-based plasmids. Plasmid 61:171–175
Ostash B, Doud EH, Lin C, Ostash I, Perlstein DL, Fuse S, Wolpert M, Kahne D, Walker S (2009) Complete characterization of seventeen-step moenomycin biosynthetic pathway. Biochemistry 48:8830–8841
Ostash I, Ostash B, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V (2008) Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol Lett 285:195–202
Sambrook J, Russel DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA (2002) The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem Biol 9:519–531
Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K (1996) Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178:7276–7284
Stinchi S, Azimonti S, Donadio S, Sosio M (2003) A gene transfer system for glycopeptide producer Nonomuraea sp. ATCC39727. FEMS Microbiol Lett 225:53–57
Sun J, Hesketh A, Bibb M (2001) Functional analysis of relA and rshA, two relA/spoT homologues of Streptomyces coelicolor A3(2). J Bacteriol 183:3488–3498
Vogtli M, Chang PC, Cohen SN (1994) afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol Microbiol 14:643–653
Taylor J, Li X, Oberthür M, Zhu W, Kahne D (2006) The total synthesis of moenomycin A. J Am Chem Soc 128:15084–15085
Yanai K, Murakami T, Bibb M (2006) Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc Natl Acad Sci USA 103:9661–9666
Yuan Y, Fuse S, Ostash B, Sliz P, Kahne D, Walker S (2008) Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem Biol 3:429–436
Acknowledgments
This work was supported by grant Bg-01F from the Ministry of Education and Science of Ukraine (to V. F.) and NIH grant AI50855 (to S. W.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Makitrynskyy, R., Rebets, Y., Ostash, B. et al. Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37, 559–566 (2010). https://doi.org/10.1007/s10295-010-0701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0701-1