Abstract
Many filamentous fungi secrete considerable quantities of enzymes including protease, cellulase and xylanase, which are of major industrial importance. Over the past few decades, many of these fungal enzymes have been isolated and their relevant genes characterised. Solid-state fermentation (SSF), an ancient technique described as a fermentation process performed on non-soluble material whereby the material acts as a physical support and as a source of nutrients, is widely employed in the production of industrially important enzymes. Control mechanisms governing gene expression in SSF however, have been rarely studied. The influence of carbon and nitrogen sources on the production and transcriptional regulation of hydrolase enzymes secreted by an Aspergillus strain was investigated with the hope of expanding on the relatively small amount of knowledge regarding cellular control of gene expression. This study involved screening a collection of fungal strains for protease, cellulase and xylanase production under SSF conditions. From this, one fungal strain was then chosen for further analysis. Factors affecting the secretion of the hydrolase enzymes were optimised, and following this, the influence of nutritional supplementation on the production and transcriptional regulation of the enzymes was investigated. Real-time PCR techniques were used to assess the relative expression levels of genes encoding hydrolase activities and of the genes encoding regulatory elements such as AreA, PacC and CreA in an effort to identify possible transcriptional regulation mechanisms. The complexity of gene regulation under SSF conditions became apparent during the study, as other factors such as post-transcriptional regulation appeared to play a far greater role than previously imagined.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past few years many genes playing fundamental roles in the regulation of gene expression have been isolated and characterised from fungal species including Aspergillus [1]. The ability of organisms to regulate gene expression is essential for their development and adaptation to environmental changes, which in turn are essential for survival [2, 3]. Despite the fact that filamentous fungi have long been exploited for the production of enzymes under SSF conditions, very little research has been documented as yet to illustrate the control mechanisms of gene expression in SSF. It has been documented that the expression of some hydrolase-producing genes are controlled at the level of transcription [4, 5]; however, events that occur after transcription and translation are much less understood in fungi and are consequently much harder to control [6]. A number of key regulatory circuits, including carbon catabolite repression (CCR) and nitrogen metabolism, have been described under submerged fermentation conditions (SmF) in Aspergilli and the main regulatory genes identified. In addition to metabolic regulation, another important regulatory phenomenon, namely in response to external pH, has also been characterised.
In this study, the influence of carbon and nitrogen sources on the production and transcriptional regulation of cellulase, xylanase and protease produced by Aspergillus oryzae M2K under solid-state conditions was investigated using real-time PCR techniques with the aim of clarifying the control mechanisms and pathways involved in nutrient metabolism. In Aspergillus species, enzyme secretion is influenced by many mechanisms that are controlled by certain key transcription factors. With this in mind the transcript accumulation levels of 12 genes were examined after cultivation of A. oryzae M2K fermentation supplemented with various carbon and nitrogen sources (ammonium phosphate, d-mannitol, yeast extract, glucose, carboxymethyl cellulose (CMC), peptone and casein). Each gene utilised in this study is explained below with regard to its role in regulation.
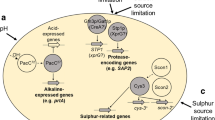
The creA gene is the main gene governing carbon CCR in Aspergillus species [3, 7, 8]. This mechanism allows fungi to cope smoothly with changes in the carbon source present in their environment. Glucose is extremely abundant in nature, and many living organisms have evolved efficient mechanisms to utilise glucose as a carbon and energy source [9]. In the presence of a readily metabolisable carbon source, such as glucose, the expression of structural genes required for utilisation of alternative carbon sources is decreased [10]. creA is known to be involved in the repression of many industrially important enzymes, including cellulase, protease and xylanase [2, 11, 12], and has also been shown to be regulated at a transcriptional and post-transcriptional level [4, 13].
With respect to nitrogen metabolism, Aspergillus species are capable of utilising a wide range of nitrogen-containing compounds as a sole nitrogen source, including ammonia, nitrate and nitrites [14, 15]. The areA gene encodes the major nitrogen regulatory protein, which activates transcription of many structural genes encoding enzymes for nitrogen source catabolism under nitrogen-limiting conditions. The availability of favoured nitrogen sources prevents expression of enzymes required for the catabolism of less preferred nitrogen sources [16–18]. In relation to enzyme regulation, the results of the role of areA in cellulase and protease regulation are still in its infancy; however, a role for areA in the expression of cellulases in A. nidulans has been suggested [19, 20], while research by Christensen et al. [17] showed that A. oryzae transformants did not form clearing halos, which is an indicator of extracellular protease activity, in the presence of ammonium. In addition to areA, a number of other genes have been identified as influencing the nitrogen regulatory circuit including tamA [21]. It has been proposed that tamA is a coactivator of areA and is therefore required for full expression of genes under the control of areA [22].
The nitrate assimilation pathway is composed of two transporters, nitrate reductase encoding the structural gene niaD and nitrite reductase encoding niiA [23, 24]. These structural genes are very closely linked and probably contiguous in Aspergillus [25]. The respective genes are subject to regulation at the level of transcription, including nitrogen metabolite repression mediated by AreA, and induction by nitrate or nitrite, mediated by the pathway-specific transcription factor NirA [25, 27]. Both transcription factors act synergistically to regulate the structural genes when nitrogen is limiting and either nitrate or nitrite is available.
In addition to adapting to metabolic changes, microorganisms must also be able to adapt physiologically to changes in environmental pH as it influences many factors including growth. Failure to do so would negatively impact on an organism’s competitiveness and therefore success [28]. The ability of fungi to thrive over a wide pH range is partly due to a genetic regulatory system that tailors gene expression to the ambient pH [29–31]. pacC is one of seven genes involved in pH regulation, the others being the pal genes. The PacC protein functions as a transcriptional activator of alkaline-expressed genes and a repressor of acid-expressed genes. When growth is at acidic pH, PacC is non-functional as a transcriptional activator of alkaline expressed genes and a repressor of acid expressed genes. Under alkaline growth conditions, the pal ambient pH signalling pathway causes the activation of the PacC protein as both an activator of alkaline-expressed genes and a repressor of acid-expressed genes. The pal/PacC pathway tailors the synthesis of extracellular hydrolytic enzymes including protease to the needs imposed by the ambient pH [32].
Fungi including A. oryzae are well known for their ability to secrete proteolytic enzymes, and as such gene expression has been studied a great deal. Analysis of the proteolytic spectrum revealed four extracellular acid proteases including pepA, which is only expressed when the preferred carbon and nitrogen source is not available to the cell [33, 34]. This extracellular protease is strictly pH regulated [35] and only expressed in solid-state fermentation (SSF) cultures [36]. In addition to pepA only being expressed in SSF, the glucoamylase gene glaB also follows this trait. While pepA is the main gene involved in protease regulation, the transcriptional activator XlnR mainly controls xylanolytic systems of A. niger [37–39]. XlnR regulates the transcription of the xlnB, xlnC and xlnD genes encoding the main xylanolytic enzymes (endoxylanases B and C and β-xylosidase, respectively) [5, 40]. Xylanolytic production is regulated by three independent mechanisms, ambient pH, carbon catabolite repression and specific induction in the presence of xylan or xylose. Not only does XlnR control transcriptional activation of the xylanolytic genes, it also directs transcription of the endoglucanase (eglA and eglB) and cellobiohydrolase (cbhA and cbhB) encoding genes, indicating that the transcriptional activation mechanism in Aspergillus is partially shared between cellulases and xylanases [41]. Finally, the brlA gene is one of three genes expressed during conidiophore development [42]. It has also been reported to directly affect enzyme production, as secretion occurs mainly from hyphal tips [43, 44].
In this study, an Aspergillus strain was isolated for further research. Enzymatic and molecular analyses were performed to further enhance the understanding of molecular mechanisms involved in enzyme regulation under SSF conditions, and the results are discussed. It was anticipated that this type of analysis would enable a direct evaluation of the effect of nutrient supplementation on gene expression.
Materials and methods
Microorganism
Several fungal strains were screened to assess their enzyme-producing capabilities under SSF (data not shown). Of these, one fungal strain, Aspergillus oryzae M2K, was chosen for further analysis. The strain was maintained on PDA slants at 4°C until required for inoculation.
Culture conditions for SSF
Initially seed culture incubations were carried out in 500-ml conical flasks containing 200 ml cornstarch medium (g/l: glucose 5, peptone 18, KCl 0.5, MgSO4·7H2O 1.5, KH2PO4 1.0 and starch from corn 60), incubated at 30°C for 72 h. A dilution (1 in 4) of the mycelium suspension was used as inoculum in all SSF experiments. A modified method of a previously described technique was used for all SSF fermentations [45].
Enzyme extraction
Following optimal incubation, enzymes were extracted using a modified method described by Tunga et al. [46].
Enzyme assays
Protease activity was determined as described previously [47] with modified conditions including the use of 5 mM glycine/HCl buffer pH 2.5 as substrate. In addition, samples were incubated for 30 min at 40°C. Previously described techniques [48] were used for determination of cellulase and xylanase activity. Modifications included the use of 50 mM sodium acetate buffer pH 4.8 as substrate for cellulase assays. Both enzyme assays used dinitro salicylic acid (DNS) as stopping solution. Following this step both sets of samples were incubated at 100°C for 5 min and then cooled in an ice bath prior to spectrophotometric analysis at λ540nm. All the above measurements were made in quadruplicate, and all enzymatic activities were expressed as units per gramme of dry weight, HUT g−1 in the case of protease, CMCU g−1 for cellulase and Xu g−1 for xylanase.
Molecular techniques
RNA extraction and reverse transcription
Quadruplicate samples of SSF cultures were snap frozen in liquid nitrogen and ground using a sterilised pestle and mortar. Total fungal RNA was isolated from using the RNAqueous® Kit according to the manufacturer’s instructions. Total RNA integrity was determined by agarose gel electrophoresis, with purity established by calculating the ratio of the absorbance readings at 260 and 280 nm and quantified using the 260-nm absorbance reading. The ratio should fall in the range of 1.8–2.1.
Reverse transcription of RNA (1 μg) was then carried out using the High Capacity cDNA Reverse Transcription Kit according to manufacturer’s instructions. The cDNA was then quantified spectrophotometrically at 260 and 280 nm, and stored at −80°C until required for real-time PCR analysis.
PCR primers
Each gene was amplified from cDNA. Primers for each chosen gene were designed based on sequences obtained from the nucleotide database at NCBI web page. Primer sequences are represented in Table 1.
Polymerase chain reaction
A typical PCR reaction contained the following: 1 μg cDNA, primers (500 ng each); MgCl2 (2.5 mM); (NH4)2SO4 reaction buffer; dNTPs (500 μM each) and sterile water to 98 μl. Aliquots (15 μl) of the reactions were analysed on a 2% (w/v) agarose gel. Amplification by PCR was performed by modified methods previously described [49].
One-step cloning reaction
PCR products were cloned in the plasmid vector pCR2.1 using the TA cloning® kit according to the manufacturer’s protocol.
Subcloning
DNA fragments were ligated to plasmid using the Rapid DNA Ligation kit, according to the manufacturer’s instructions. The ligation reaction was carried out at room temperature using a T4 DNA ligase.
Transformation of competent cells
Escherichia coli strain INVαF′ competent cells were transformed with insert-containing vectors according to the manufacturer’s instructions. The pCR 2.1™ vector contains an ampicillin resistant gene used in the selection and maintenance of E. coli. INVαF E.coli cells do not express the lac repressor, which represses transcription from the lac promoter. The lac promoter is responsible for bacterial expression of the lacZα fragment that allows blue white screening of putative transformants to be performed in the presence of X-Gal. Cells transformed with the insert-containing plasmid appear white due to the interruption of the reading frame of the lacZ gene and are referred to as lacZ disruptants. Those cells, which have not taken up the plasmid-containing insert, appear blue because of β-galactosidase activity in the presence of X-Gal as the lacZ gene is not interrupted [50].
Restriction enzyme digestion of DNA
All restriction enzyme digests of DNA were carried out under optimal reaction conditions as specified by the manufacturer of the restriction enzyme.
Sequencing of PCR products
Sequencing of insert DNA was carried out by Cogenics (UK) using M13 forward and reverse primers. Sequence and homology data were analysed using the Basic Local Alignment Search Tool (BLAST) online at the National Centre for Biotechnology Information (NCBI) homepage.
Real-time PCR primer sequences
PCR-amplified gene fragments were used as template in the design of real-time PCR primer sets. The real-time PCR primers were designed using Primer Express Software (Version 2.0, Applied Biosystems, Foster City, CA). The primers were commercially prepared by Sigma-Genosys Inc. The BLAST tool was used to confirm the homology of primer sets. Primer sequences are displayed in Table 2.
Quantitative real-time PCR assay
All quantitative real-time PCR amplifications were carried out using the Applied Biosystems 7500 Fast Real-Time PCR system (Foster City, CA) in accordance with an experimental procedure previously described by Liu and Saint [51]. Following amplification, the cycle threshold (C T) values of the target and endogenous control genes were transformed to raw quantities according to the 2−ΔΔCT method for relative quantification [52]. Each of the cDNA samples was treated separately, and the results after the 2−ΔΔCT calculation were averaged. Results (2−ΔΔCT) were plotted as log10 transformations.
Assessment of the specificity of real-time PCR primers
The specificities of the primer sets utilised for the real-time PCR amplification were evaluated by performing melting curve analysis [53, 54].
Assessment of endogenous control efficiency
The effect of treatment on the expression of the endogenous control genes was examined in order to validate the endogenous control gene for use in the quantitative real-time PCR assay. Following the analysis of a number of common reference genes [55–57] (GAPDH, β-actin and β-tubulin), the β-tubulin gene was selected as the endogenous control gene because of its stability across all samples analysed regardless of treatment.
Assessment of real-time PCR assay efficiency
Prior to gene expression analysis, the real-time primers were assessed for accuracy by confirming that the amplification efficiencies of the target genes (Table 2) and the endogenous control gene (β-tubulin) were approximately equal. The cDNAs were diluted over a 100-fold range and amplified by real-time PCR using gene specific primers. A method previously described was used to determine efficiency of the reactions [25, 58]. Validation of the 2-ΔΔCT method is represented in Fig. 1.
Statistical analysis
Data were analysed using the Minitab statistical software package version 15.0 (Coventry, UK). Analysis programmes used included Pearson’s correlation and ANOVA.
Effect of carbon and nitrogen supplementation on gene expression
Subsequent to real-time PCR amplification, the cycle threshold (CT) number was extracted for both reference gene and target gene with auto baseline and manual threshold. The concentration of supplemental carbon and nitrogen sources was then plotted against the log10 relative quantification value and enzyme activity (% RA), which allowed for the direct comparison of the effect of nutritional supplementation on both gene expression and activity. Data were analysed using one-way analysis of variance (ANOVA). A significance level of 95% (P < 0.05) was used to discriminate between significant and non-significant increases or decreases in enzyme activity or gene expression. In addition, an evaluation was made of the strength and direction of the association using Pearson’s correlation coefficient. The measured correlation coefficient is always between −1 and +1; the closer the correlation is to ±1, the closer the relationship is to being perfectly linear.
Results
Effect of glucose supplementation on transcriptional regulation of enzyme production
The effect of glucose supplementation on transcriptional regulation of enzyme production by A. oryzae M2K is represented in Fig. 2. In terms of enzymatic productivity, glucose addition had no effect on cellulase activity, and only a slight decrease in xylanase activity was evident. It was also apparent that there was no association with the corresponding gene transcripts, xlnR and eglA, respectively. Statistical analysis (ANOVA) of protease activity revealed that there was a significant (P < 0.05) decrease in protease activity at the 10% (w/v) supplemental level, despite the expression of the protease gene, pepA, increasing at all inclusion levels. Glucose supplementation positively influenced areA expression with a significant (P < 0.05) increase noted at all inclusion levels relative to the unsupplemented control, while tamA expression significantly increased at 1 and 5% inclusion levels; however, it remained downregulated. The pH regulatory genes, pacC and palA, were in a derepressed state; however, palA expression positively increased at an inclusion level of 10% (w/v). brlA expression significantly (P < 0.05) increased at all inclusion levels when compared to the unsupplemented control; however, it remained downregulated. Overall, the strain appeared to move into a carbon-repressed state evidenced by an increase in creA transcription with increasing glucose concentration.
Analysis of gene expression results was also performed using Pearson’s correlation. It must be stressed that this type of statistical approach only measures the relationship between two variables and does not allow for determination of cause and effect. With respect to gene associations, areA, brlA, palA, pepA, pacC, creA, tamA and xlnR were all positively correlated with each other (P < 0.05), but negatively with pH (P < 0.05). In summary, the addition of glucose resulted in all the transcription factors being downregulated relative to the unsupplemented control.
Effect of d-mannitol supplementation on transcriptional regulation of enzyme production
There was a statistically significant (P < 0.05) linear decrease in cellulase activity with increasing d-mannitol concentration (Fig. 3). Both xylanase and protease activity remained relatively constant at 1 and 5% inclusion levels; however, a significant (P < 0.05) decrease in enzyme activity was noted at 10% inclusion relative to d-mannitol addition. Despite a linear decrease in cellulase activity, one-way statistical analysis revealed that eglA gene expression was significantly (P < 0.05) upregulated with increasing d-mannitol concentration when compared to the control. In addition, there was no apparent relationship between xylanase activity and xlnR gene expression as the xlnR gene significantly increased at each inclusion level despite a trend towards a decrease in activity levels. The protease encoding gene (pepA) also appeared to have no association with the measured proteolytic activity, thus suggesting the involvement of other elements. Overall it was apparent that there were distinct differences noted with respect to gene expression, with most genes being upregulated following d-mannitol supplementation. The organism was in a carbon-repressed state as evidenced by the upregulation of creA at 5 and 10% (w/v).
Analysis using ANOVA also showed that the regulation of a number of genes was significantly (P < 0.05) different (both up- and downregulated) when compared to the control at all inclusion levels. These included hapB, palA, areA, brlA, niaD, glaB, eglA and xlnR. In terms of associations, Pearson’s analysis indicated that tamA gene expression was negatively correlated with protease activity (P < 0.05), although there was no noted effect on areA.
Effect of CMC supplementation on transcriptional regulation of enzyme production
From Fig. 4 it was clear that the addition of CMC caused an overall decrease in all hydrolytic enzyme activities with increasing concentration. Statistical analysis revealed a significant (P < 0.05) difference at 3% (w/v) inclusion levels for all enzymatic activities when compared with their unsupplemented controls. The protease encoding transcript (pepA) was upregulated at 1, 2 and 3% (w/v) despite a decrease in protease activity. One-way analysis of variance indicated that a number of genes were significantly (P < 0.05) upregulated at all supplemental concentrations, including hapB, pepA, brlA, tamA, pacC, creA and glaB. CMC addition also caused the downregulation of niaD, eglA and xlnR. Despite differing CMC concentrations, the expression of the creA gene remained relatively constant. In essence, the strain remained in a carbon-repressed state relative to the unsupplemented control. Pearson’s analysis indicated that expression of a number of genes encoding regulatory elements, namely areA, palA and tamA, were negatively associated (P < 0.05) with protease, cellulase and xylanase activity. Cellulase and xylanase activity were negatively correlated (P < 0.05) with areA, palA, pacC, tamA and eglA. Gene associations suggested a positive relationship (P < 0.05) among pacC, areA, palA and pepA.
Effect of ammonium phosphate supplementation on transcriptional regulation of enzyme production
In the presence of ammonium phosphate, there was an inverse relationship between cellulase and protease activity (Fig. 5). With respect to protease activity, there was a strong linear decrease with a significant difference (P < 0.05) recorded at 1 and 10% inclusion levels when compared to the unsupplemented control. Cellulase activity increased slightly from 1 to 5% and remained relatively constant at a 10% (w/v) inclusion level. Xylanase activity was statistically different (P < 0.05) to the control at all supplementation levels. It is also apparent that the inclusion of ammonium phosphate caused the upregulation of the majority of genes.
Statistical analysis using one-way ANOVA revealed that a number of genes were significantly (P < 0.05) increased at all additions, including hapB, palA, areA, brlA, niaD, glaB and tamA. In addition, nutrient supplementation resulted in an increase in pacC and creA gene expression with increasing ammonium phosphate concentration, effecting a change from a derepressed to a repressed state. In terms of associations, protease and cellulase activity were positively correlated (P < 0.05) with their associated gene transcripts. This was the first instance noted in this study where enzyme activity could be correlated with the corresponding gene. In terms of relationships among regulatory elements, areA was positively correlated (P < 0.05) with hapB, palA, xlnR, niaD and glaB, while negatively (P < 0.05) with pepA and protease activity. It has been documented that areA is involved in controlling the expression of some of these transcription factors [25, 26, 33]. palA was negatively correlated with pacC and creA, while pacC was positively associated with creA, tamA and eglA. In terms of pH, Pearson’s analysis indicated that negative correlations (P < 0.05) were noted with brlA, pacC, creA and tamA, while a positive association was noted with eglA and xylanase activity. In addition, pH was clearly associated with protease activity and pepA expression (P < 0.05), as evidenced by the decrease noted with increasing ammonium phosphate concentration. Overall, the strain is in a nitrogen-repressed state, evidenced by the upregulation of areA gene expression.
Effect of casein supplementation on transcriptional regulation of enzyme production
As can be noted in Fig. 6, the majority of genes assessed were upregulated following casein addition. With respect to enzyme activity, protease remained at a relatively constant level despite an increase in casein concentration. In addition, the protease-encoding transcript (pepA) was upregulated with increasing casein concentration and therefore had no association with the measured enzyme activity. Similarly, cellulase activity decreased despite a dramatic upregulation in eglA gene expression with increasing casein concentration. Xylanase activity remained relatively constant, despite an increase in the expression of the xlnR regulatory factor being observed at the 5 and 10% (w/v) inclusion levels, once again implying the involvement of additional control mechanisms. Statistical analysis (ANOVA) showed that areA, tamA, pepA and eglA genes were positively influenced (P < 0.05) by increasing casein concentration, whereas brlA, pacC, creA and glaB expression levels significantly (P < 0.05) decreased.
In terms of gene associations, Pearson’s correlation indicated a number of relationships. areA was negatively (P < 0.05) correlated with brlA, palA, pacC, creA and glaB, but positively with pepA, tamA, eglA and xlnR. pacC was negatively associated with pepA, but positively with creA. In terms of pH, there was a negative association with pacC and creA and a positive one with tamA and eglA. Overall, with increasing casein concentration, the strain moved from a nitrogen-derepressed to -repressed state as evidenced by an increase in areA gene expression with increasing concentration.
Effect of peptone supplementation on transcriptional regulation of enzyme production
The effect of peptone supplementation on transcriptional regulation of enzyme production is represented in Fig 7. In terms of enzyme activity, protease activity was significantly (P < 0.05) downregulated with increasing peptone concentration despite an increase in pepA gene expression. Xylanase activity increased significantly (P < 0.05) at all inclusion levels, while expression of the regulatory factor xlnR increased from 1 to 5% (w/v); however, it did drop slightly at 10% (w/v). Cellulase activity remained relatively stable despite the upregulation of eglA gene expression with increasing concentration. Statistical analysis (ANOVA) revealed that the majority of genes were significantly (P < 0.05) enhanced at all additions (1, 5 and 10%) with increasing nutrient concentration. These included palA, pepA, areA, brlA, tamA and eglA. In addition pacC expression increased with increasing peptone concentration and was significantly (P < 0.05) upregulated at the 10% (w/v) inclusion level. Peptone addition at 1% (w/v) effected a downregulation of the majority of regulatory elements, while increasing nutrient concentrations to 5 and 10% (w/v) reversed this trend. The upregulation of areA at 5 and 10% (w/v) would indicate that the organism was in a nitrogen-repressed state. Overall, the addition of peptone resulted in a reduction in protease activity, and potentially there may be a very complex regulatory mechanism at work as pepA transcription was increased but protease activity was decreased.
Effect of yeast extract supplementation on transcriptional regulation of enzyme production
Yeast extract addition brought about a downregulation of the majority of genes at each supplemental level (Fig. 8). An inverse relationship between xylanase and protease activity was apparent (P < 0.05). In addition, there was a trend towards increasing xlnR expression with increasing activity, indicating that xylanase activity was regulated by its corresponding activator at the level of transcription. However, pepA gene expression was downregulated. Cellulase production remained constant despite varying supplement concentrations, and in addition, no relationship with eglA gene expression was noted.
Statistical analysis revealed that there was a significant increase in the expression of a number of regulatory elements with increasing nutrient concentration. These included areA, brlA, tamA, pacC and creA. In terms of gene associations, pH was positively correlated (P < 0.05) with numerous genes, including brlA, palA, pepA, pacC, creA, tamA, xlnR and xylanase activity. Protease activity had negative associations with areA, brlA, palA, pepA, pacC, creA and tamA genes, and due to the inverse relationship, all these factors were positively correlated with xylanase production. In simple terms, in the presence of yeast extract, A. oryzae M2K produced protease at the expense of xylanase, and cellulase activity remained unaffected. Overall, the organism remained in a carbon-derepressed and nitrogen-derepressed state as evidenced by the downregulation of creA and areA, respectively, compared to the unsupplemented control.
Discussion
Over the last decade, research has focussed mainly on gene regulation in submerged fermentation (SmF); however, with current production interests focussing more heavily on SSF, intense efforts are being made to expand on the relatively small amount of knowledge regarding cellular control of gene expression [48]. In this study, the expression levels of 12 transcript genes were examined after the cultivation under SSF conditions. Following real-time PCR analysis, it became clear that the mechanisms involved in transcriptional regulation were quite complex. Gene expression analysis revealed that noticeable differences existed between observed data generated during the course of this work and the published research detailing control in submerged fermentation.
Filamentous fungi can grow under varying environmental conditions and can utilise a wide variety of nutrients including carbon, nitrogen and sulphur sources, and it is widely known that these nutritional factors along with environmental factors such as pH can affect enzyme secretion [1]. Although the dominant factor in regulating these parameters has been documented to be controlled at the level of transcription, it became apparent in this study that it is often not the main step involved in protein expression and secretion in a fungal system. The regulation of gene expression begins at the level of transcription of DNA into mRNA [59]. The mRNA content of the cell will change as different genes are expressed in response to external factors. Although genes expressed within a cell are an indication of the proteins being produced, they may not necessarily reflect protein function. During the passage of protein secretion, proteins are subjected to various post-translational modifications, which play a crucial role in protein structure, stability and function. Major modifications can include the formation of disulphide bridges, proteolytic cleavage and glycosylation [43, 60]. Recent studies have led to the identification of over 300 post-translational modifications, and in addition many of these are regulatory and reversible [61]. However, an understanding of these regulatory mechanisms is still in its infancy in solid-state cultivation.
In terms of this study, it was apparent that there was no association between the majority of enzyme activities and their corresponding gene transcripts following nutrient supplementation. The only exception to this was following ammonium phosphate addition whereby all enzymatic activities could be positively correlated with the associated gene. It is understood that an increase or decrease in transcription does not necessarily result in an increase or decrease of the corresponding protein encoded by that gene [62]. Therefore, in this case, other mechanisms of regulation were potentially highly influential on protein production.
In addition to analysing gene transcripts and their corresponding hydrolase activities, statistical analysis including Pearson’s correlation and ANOVA (analysis of variance) were also used and produced results not commonly associated with SmF. It has been documented that the production of protease activity in SmF is regulated by pH, which is mediated by the pacC/palA signalling pathway [63]. However, following glucose supplementation there was a decrease in protease activity from 5% (w/v) to 10% (w/v) despite an increase in the pH regulated transcripts, namely pepA, pacC and palA. In terms of associations following d-mannitol supplementation, Pearson’s analysis indicated that tamA gene expression was negatively correlated with protease activity (P < 0.05), although there was no noted effect on areA, which would have been anticipated as tamA has been suggested as a coactivator of areA [22]. These findings again indicate the complexity of the mechanisms governing transcriptional regulation in SSF. In terms of CMC addition, derivatives of cellulose including carboxymethyl cellulose (CMC) have been widely documented as a sufficient carbon source for the production of cellulase enzymes and have also been reported to induce cellulase genes [64]. Pearson’s analysis indicated that cellulase and xylanase activity were negatively correlated (P < 0.05) with areA, palA, pacC, tamA and eglA. This may imply that the main contributors towards controlling cellulase and xylanase activity were the effect of the areA/tamA interaction and pH. In Aspergillus, secreted cellulases are affected by both the carbon and nitrogen source present in the medium and therefore are regulated directly and/or indirectly by the carbon metabolism regulator CreA and the global nitrogen metabolism regulator, AreA. The putative regulatory regions of the genes controlling cellulase and xylanase activity contain potential binding sites for creA and areA; therefore, in this case areA could have had more of an influence on enzyme activity [19]. Obviously this would require further studies to validate this hypothesis. In addition, following CMC supplementation protease activity decreased despite an upregulation of pepA implying the involvement of other mechanisms not included in this study. The pepA gene is the main gene involved in the regulation of extracellular protease activity; however, in this case an upregulation in transcription levels did not result in an increase in enzymatic activity. Positive gene associations outlined in the “Effect of CMC supplementation on transcriptional regulation of enzyme production” section may imply that pH played a role in stimulating transcription of areA and tamA, thus negatively affecting protease production. However, given that the pepA transcript was upregulated relative to the unsupplemented control, it is unlikely that areA and tamA negatively affected protease activity, as their affects are known only at a transcriptional level. Other elements involved in post-transcriptional and translational control must clearly be involved.
Despite the positive relationship between protein production and gene transcript expression following ammonium phosphate addition, statistical analysis revealed results that were not anticipated, including niaD not being downregulated. niaD is a structural gene involved in the nitrate assimilation pathway and is subject to repression by nitrogen metabolism in the presence of the preferred nitrogen source such as ammonium; it is generally only expressed when the less favoured source is available [65]. In addition, it has recently been shown that niaD can become very unstable in the presence of ammonia or glutamine [66].
Following casein supplementation cellulase activity decreased dramatically despite an upregulation in eglA gene expression. This may have been influenced by XlnR as mechanisms involving this transcription factor are shared among cellulases and xylanases in Aspergillus. It is therefore possible that xlnR affected cellulase production [41]. It is also worth noting that the shared relationship between cellulase and xylanase strengthens the hypothesis that there is an evolutionary analogy between the regulation of these enzyme systems [67]. It has also been shown that casein as a nitrogen source is a sufficient inducer of proteolytic activity in submerged fermentation [47]; however, this did not appear to be the case in this instance because of the reduction in protease levels with increasing concentration. In relation to yeast extract supplementation is a mixed carbon and nitrogen source and therefore may effect transcriptional regulation in a manner different than only carbon or nitrogen supplementation. This again underlines the complexity of growth in solid-state fermentation and more importantly how nutritional supplementation may not always be beneficial in terms of enzyme productivity.
Conclusion
In summary, this study revealed the limited knowledge surrounding gene regulation of hydrolase enzymes under SSF conditions. The conditions for growth of A. oryzae differ between SSF and SmF in many aspects, including gradients of nutrients, oxygen, temperature and water availability, and consequently differences in transcriptional regulation must be expected [68]. Although this research did show that the regulation of some genes could be influenced by carbon and nitrogen addition, a full understanding of the mechanisms involved remains unclear. Throughout this research, mechanisms involved subsequent to transcription appeared to play a far greater role in regulating enzyme production than were previously detailed. No clear link can be made between the transcriptional elements involved in regulation and the noted enzyme activities even though in submerged fermentation these elements have been shown to directly regulate enzyme activity [5, 19, 69]. To develop a more comprehensive understanding of the mechanisms involved in transcriptional regulation of SSF processes, it would be necessary to consider not only transcriptional but also translational and post-translational regulation. Clearly from this work the latter two mechanisms must play a far greater role in enzyme regulation than noted in classical submerged fermentation.
In the past, mechanisms for studying gene regulation in SSF have been hampered as agri-industrial substrates such as wheat bran, rice bran, silage, corn and apple pomace contain plant polyphenols and polysaccharides, which are known to inhibit PCR and reverse transcription (RT)-PCR [70]. Currently, tremendous strides are being made in our understanding of gene regulation largely because of the powerful combination of genetics, biochemistry and molecular approaches and the isolation and characterisation of regulatory and structural genes and their protein. The insight into the mechanisms controlling the regulatory circuits will aid in SSF process design and optimisation in areas such as strain improvement, solid substrate engineering and process control. In addition, further understanding will enhance protein secretion levels in important industrial species such as Aspergillus using solid-state fermentation conditions. Thus, over the next few years the perspective that SSF will gain in significance in the industrial production of enzymes among other important products is warranted.
References
Tudzynski P, Tudzynski B (1997) Novel techniques and regulatory circuits. In: Anke (ed) Fungal biotechnology. Chapman and Hall, Weinheim, pp 229–249
Ward OP, Qin WM, Dhanjoon J, Ye J, Singh A (2006) Physiology and biotechnology of Aspergillus. Adv Appl Microbiol 58:1–75
Jacobs M, Stahl U (1995) Gene regulation in mycelial fungi. In: Kuck (ed) The mycota II: genetics and biotechnology. Springer, Berlin, pp 115–167
Lubertozzi D, Keasling JD (2009) Developing Aspergillus as a host for heterologous expression. Biotechnol Adv 27:53–75
Tamayo EN, Villanueva A, Hasper AA, Graaff LH, Ramón D, Orejas M (2008) CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol 45:984–993
Jarai G (1997) Heterologous gene expression in filamentous fungi. In: Anke (ed) Fungal biotechnology. Chapman and Hall, Weinheim, pp 251–261
Fillinger S, Panozzo C, Mathieu M, Felenbok B (1995) The basal level of transcription of the alc genes in the ethanol regulon in Aspergillus nidulans is controlled both by the specific transactivator AlcR and the general carbon catabolite repressor CreA. FEBS Lett 368:547–550
Petersen KL, Lehmbeck J, Christensen T (1999) A new transcriptional activator for amylase genes in Aspergillus. Mol Gen Genet 262:668–676
Mogensen J, Nielsen HB, Hofmann G, Nielsen J (2006) Transcription analysis using high-density micro-arrays of Aspergillus nidulans wild-type and creA mutant during growth on glucose or ethanol. Fungal Genet Biol 43:593–603
Ilmen M, Thrane C, Penttila M (1996) The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet 251:451–460
Mach RL, Strauss J, Zeilinger S, Schindler M, Kubicek CP (1996) Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol Microbiol 21:1273–1281
Katz M, Bernardo S, Cheetham B (2008) The interaction of induction, repression and starvation in the regulation of extracellular proteases in Aspergillus nidulans: evidence for a role for CreA in the response to carbon starvation. Curr Genet 54:47–55
Archer DB (2000) Filamentous fungi as microbial cell factories for food use. Curr Opin Biotechnol 11:478–483
Orejas M, Espeso EA, Tilburn J, Sarkar S, Arst MA, Penalva HNJ (1995) Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev 9:1622–1632
Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, Davies RW, Arst HNJ (1990) The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J 9:1355–1364
Peters DG, Caddick MX (1994) Direct analysis of native and chimeric GATA specific DNA binding proteins from Aspergillus nidulans. Nucleic Acids Res 22:5164–5172
Christensen T, Hynes MJ, Davis MA (1998) Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl Environ Microbiol 64:3232–3237
Wilson RA, Arst HN (1998) Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “Streetwise” GATA family of transcription factors. Microbiol Mol Biol Rev 62:586–596
Lockington RA, Rodbourn L, Barnett S, Carter CJ, Kelly JM (2002) Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet Biol 37:190–196
Aro N, Pakula T, Penttila M (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev 29:719–739
Davis MA, Small AJ, Kourambas S, Hynes MJ (1996) The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J Bacteriol 178:3406–3409
Morozov IY, Galbis-Martinez M, Jones MG, Caddick MX (2001) Characterization of nitrogen metabolite signalling in Aspergillus via the regulated degradation of areA mRNA. Mol Microbiol 42:269–277
Takasaki K, Shoun H, Nakamura A, Hoshino T, Takaya N (2004) Unusual transcription regulation of the niaD gene under anaerobic conditions supporting fungal ammonia fermentation. Biosci Biotechnol Biochem 68:978–980
Caddick MX, Jones MG, van Tonder JM, Le Cordier H, Narendja F, Strauss J, Morozov IY (2006) Opposing signals differentially regulate transcript stability in Aspergillus nidulans. Mol Microbiol 62:509–519
Marzluf GA (1981) Regulation of nitrogen metabolism and gene expression in fungi. Microbiol Rev 45:437–461
Burger G, Strauss J, Scazzocchio C, Lang BF (1991) nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. J Mol Cell Biol 11:5746–5755
Muro-Pastor MI, Gonzalez R, Strauss J, Narendja F, Scazzocchio C (1999) The GATA factor of AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J 18:1584–1597
Nozawa SR, Ferreira-Nozawa MS, Martinez-Rossi NM, Rossi A (2003) The pH-induced glycosylation of secreted phosphatases is mediated in Aspergillus nidulans by the regulatory gene pacC-dependent pathway. Fung Genet Biol 39:286–295
Mingot JM, Espeso EA, Díez E, Peñalva MÁ (2001) Ambient pH signaling regulates nuclear localization of the Aspergillus nidulans PacC transcription factor. Mol Cell Biol 21:1688–1699
Diez E, Alvaro J, Espeso EA, Rainbow J, Suarez T, Tilburn J, Arst HN, Penalva MA (2002) Activation of the Aspergillus PacC zinc finger transcription factor requires two preteolytic steps. EMBO J 21:1350–1359
Arst HN, Penalva MA (2003) pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet 19:224–231
Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Munera-Huertas T, Stanton L, Hervas-Aguilar A, Espeso EA, Tilburn J, Arst HN Jr, Penalva MA (2007) Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell 6:2365–2375
van Kuyk PA, Cheetham BF, Katz ME (2000) Analysis of two Aspergillus nidulans genes encoding extracellular proteases. Fung Genet Biol 29:201–210
Hoffman B, Breuil C (2003) Analysis of the distribution and regulation of three representative subtilase genes in sapstaining fungi. Fung Genet Biol 41:274–283
van den Hombergh JPTW, van de Vondervoort PJI, Fraissinet-Tachet L, Visser J (1997) Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol 15:256–263
Iwashita K (2002) Recent studies of protein secretion by filamentous fungi. J Biosci Bioeng 94:530–535
Hasper AA, Visser J, de Graaff LH (2000) The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression. J Mol Microbiol 36:193–200
Endo Y, Yokoyama M, Morimoto M, Shirai K, Chikamatsu G, Kato N, Tsukagoshi N, Kato M, Kobayashi T (2008) Novel promoter sequence required for inductive expression of the Aspergillus nidulans endoglucanase gene eglA. Biosci Biotechnol Biochem 72:312–320
Stricker A, Mach R, de Graaff L (2008) Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl Microbiol Biotechnol 78:211–220
Pérez-González J, van Peij N, Bezoen A, de Graaff LH (1998) Molecular cloning and transcriptional regulation of the Aspergillus nidulans xlnD gene encoding a β-xylosidase. Appl Environ Microbiol 64:1412–1419
Marui J, Tanaka A, Mimura S, de Graaff LH, Visser J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N (2002) A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet Biol 35:157–169
Han S, Navarro J, Greve RA, Adams TH (1993) Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J 12:2449–2457
Radzio R, Kück U (1997) Synthesis of biotechnologically relevant heterologous proteins in filamentous fungi. Proc Biochem 32:529–539
te Biesebeke R, Record E, van Biezen N, Heerikhuisen M, Franken A, Punt P, van den Hondel C (2005) Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Appl Microbiol Biotechnol 69:44–50
Ellaiah P, Adinarayana K, Bhavani Y, Padmaja P, Srinivasulu B (2002) Optimization of process parameters for glucoamylase production under solid state fermentation by a newly isolated Aspergillus species. Proc Chem 38:615–620
Tunga R, Shrivastava B, Banerjee R (2003) Purification and characterization of a protease from solid state culture of Aspergillus parasiticus. Proc Biochem 38:1553–1558
Tremacoldi CR, Watanabe NK, Carmona EC (2004) Production of extracellular acid protease by Aspergillus clavatus. W J Microbiol Biotechnol 20:639–642
Botella C, de Ory I, Webb C, Cantero D, Blandino A (2005) Hydrolytic enzyme production by Aspergillus awamori on grape pomace. Biochem Eng J 26:100–106
Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction products by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus. Proc Natl Acad Sci USA 88:7276–7280
Chaffin DO, Rubens CE (1998) Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91–99
Liu W, Saint DA (2002) Validation of a quantitative method for real time PCR kinetics. Biochem Biophys Res Commun 294:347–353
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM, Moorman AFM (2002) Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green 1 depends on cDNA synthesis conditions. Anal Biochem 307:63–69
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, Stahlberg A, Zoric N (2006) The real-time polymerase chain reaction. Mol Aspect Med 27:95–125
Thellin O, Zorzi W, Lakaye B, de Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Sweeney MJ, Pamies P, Dobson ADW (2000) The use of reverse transcription-polymerase chain reaction (RT-PCR) for monitoring aflatoxin production in Aspergillus parasiticus 439. Int J Food Microbiol 56:97–103
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313:856–862
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–512
Campbell NA, Reece JB, Mitchell LG (1999) From gene to protein. In: Biology, 5th edn. Benjamin-Cummings, CA, pp 294–318
Nevalainen KMH, Te’o VSJ, Bergquist PL (2005) Heterologous protein expression in filamentous fungi. Trend Biotechnol 23:468–474
Bhadauria V, Zhao WS, Wang LX, Zhang Y, Liu JH, Yang J, Kong LA, Peng YL (2007) Advances in fungal proteomics. Microbiol Res 162:193–200
Brunner K, Lichtenauer A, Kratochwill K, Delic M, Mach R (2007) Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr Genet 52:213–220
Espeso EA, Roncal T, Díez E, Rainbow L, Bignell E, Álvaro J, Suárez T, Denison SH, Tilburn J, Arst HNJ, Peñalva MA (2000) On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J 19:719–728
Chikamatsu G, Shirai K, Kato M, Kobayashi T, Tsukagoshi N (1999) Structure and expression properties of the endo-β-1, 4-glucanase A gene from the filamentous fungus Aspergillus nidulans. FEMS Microbiol Lett 175:239–245
Chang PK, Ehrlich KC, Linz JE, Bhatnager D, Cleveland TE, Bennett JW (1996) Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr Genet 30:68–75
Olszewska A, Król K, Weglenski P, Dzikowska A (2007) Arginine catabolism in Aspergillus nidulans is regulated by the rrmA gene coding for the RNA-binding protein. Fungal Genet Biol 44:1285–1297
Oda K, Kakizono D, Yamada O, Lefuji H, Akita O, Iwashita K (2006) Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl Environ Microbiol 72:3448–3457
te Biesebeke R, van Biezen N, de Vos WM, van den Hondel CAMJJ, Punt PJ (2005) Different control mechanisms regulate glucoamylase and protease gene transcription in Aspergillus oryzae in solid-state and submerged fermentation. Appl Microbiol Biotechnol 67:75–82
MacCabe AP, Orejas M, Tamayo EN, Villanueva A, Ramón D (2002) Improving extracellular production of food-use enzymes from Aspergillus nidulans. J Biotechnol 98:43–54
Cohen R, Yarden O, Hadar Y (2002) Lignocellulose affects Mn2+ regulation of peroxidase transcriptional levels in solid state cultures of Pleurotus ostreatus. Appl Environ Microbiol 68:3156–3158
Acknowledgments
The financial support of Alltech Inc. is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McKelvey, S.M., Murphy, R.A. Analysis of wide-domain transcriptional regulation in solid-state cultures of Aspergillus oryzae . J Ind Microbiol Biotechnol 37, 455–469 (2010). https://doi.org/10.1007/s10295-010-0691-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0691-z