Abstract
l-Valine biosynthesis was analysed by comparing different plasmids in pyruvate-dehydrogenase-deficient Corynebacterium glutamicum strains in order to achieve an optimal production strain. The plasmids contained different combinations of the genes ilvBNCDE encoding for the l-valine forming pathway. It was shown that overexpression of the ilvBN genes encoding acetolactate synthase is obligatory for efficient pyruvate conversion and to prevent l-alanine as a by-product. In contrast to earlier studies, overexpression of ilvE encoding transaminase B is favourable in pyruvate-dehydrogenase-negative strains. Its amplification enhanced l-valine formation and avoided extra- and intracellular accumulation of ketoisovalerate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
White biotechnology is a prospering field, as it deals with industrial production of substances used for food, feed and fuel, or other chemical conversions [1]. Amino acids are important products in the area of white biotechnology, with a market value of USD 4.5 billion in 2004 [2]. Besides their major use for food (l-glutamate) and feed (l-lysine, l-tryptophan, l-threonine), amino acids are also used for pharmaceutical purposes. For example, l-valine is an essential amino acid used for total artificial nutrition and for synthesis of antiviral drugs [3]. Hence, this amino acid is a target for development of a microbial production process.
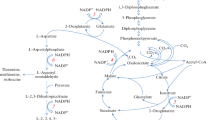
Biochemically, l-valine is produced from two molecules of pyruvate (Fig. 1). These are condensed by acetolactate synthase (AHAS, encoded by the genes ilvBN) to acetolactate [4]. Acetolactate is reduced and isomerised to dihydroxyisovalerate (DHIV) by acetolactate isomeroreductase (AHAIR, ilvC gene product). DHIV is further dehydrated to ketoisovalerate (KIV), catalysed by dehydroxy acid dehydratase (DHAD, ilvD gene product). The main transaminase for the transamination of KIV is transaminase B, encoded by the ilvE gene. It uses glutamate as an amino donor to form l-valine. l-Valine can be exported by the branched-chain amino acid exporter (brnFE gene product) and can also pass the cell membrane by diffusion [5]. The branched-chain amino acids can be imported from the medium by the specific carrier system BrnQ [6].
l–Valine pathway and selected reactions from pyruvate in C. glutamicum. Enzymes are shown in grey ellipses and genes in black boxes. PDHC pyruvate dehydrogenase complex, PQO pyruvate-quinone oxidoreductase, CS citrate synthase, AHAS acetolactate synthase, AHAIR acetohydroxy acid isomeroreductase, DHAD dihydroxy acid dehydratase, TAB transaminase B, BrnFE branched-chain amino acid exporter, DHIV dihydroxyisovalerate, KIV ketoisovalerate, CoA coenzyme A
Notwithstanding some early approaches beginning in the 1950s using a wide range of organisms such as Micrococcus or Aerobacter [7, 8], l-valine formation has mainly been analysed using C. glutamicum strains [9]. An alternative approach using a modified E. coli strain was presented lately [10]. Recently, several approaches have been published for the development of a fermentation process for l-valine formation; they are based on sufficient precursor availability and optimal precursor conversion towards the desired product: Enhanced pyruvate precursor availability can be achieved by pantothenate limitation [11, 12] or inactivation of the pyruvate dehydrogenase complex (PDHC) [3]. The optimisation of cofactor supply, i.e. nicotinamide adenine dinucleotide phosphate (NADPH), was shown to be another target for strain optimisation [13, 14].
So far, the optimisation of l-valine formation has mainly been based exclusively on the information gained from extracellular metabolite measurements. The optimised pyruvate supply in PDHC-deficient strains [14] is a successful example of how effective intracellular data can contribute to optimisation purposes. As part of the work presented here, we analysed the effect of different plasmids harbouring different genes of the l-valine pathway. Different combinations of l-valine biosynthesis genes were overexpressed in C. glutamicum in order to channel the carbon flux towards the desired product. Radmacher et al. [12] showed overexpression of the genes ilvBNCD to be optimal for l-valine formation. Elisakova et al. [15] were most successful with overexpression of ilvBNC in a feedback-resistant strain. After PDHC inactivation, the overexpression of the genes ilvBNCE was used for optimised l-valine production [3, 14].
In the work reported herein, we analysed cytoplasmic metabolite concentrations in PDHC-deficient C. glutamicum strains harbouring different plasmids with various combinations of genes encoding enzymes involved in l-valine biosynthesis. This was done to identify the optimal production plasmid for l-valine hyperproduction. Especially the prevention of by-product formation (e.g. pyruvate and l-alanine) attracted our attention, since the separation of l-valine and l-alanine during downstream processing is rather difficult and can make a real difference to the economics of the overall production process [9].
Materials and methods
Bacterial strains and cultivation
The wild-type C. glutamicum ATCC 13032 and the l-valine production strains C. glutamicum ΔaceE [16], C. glutamicum ΔaceE pJC4ilvBNC, C. glutamicum ΔaceE pJC4ilvBNCD, C. glutamicum ΔaceE pJC4ilvBNCE (all [14]), C. glutamicum ΔaceE pJC4ilvCDE (this work) and C. glutamicum ΔaceE ilvN CH pJC4ilvCDE (feedback-resistant allele provided by Evonik Degussa GmbH) were used in order to compare product and by-product formation and to analyse the intracellular concentrations of intermediates of l-valine biosynthesis. The feedback-resistant allele ilvN CH was obtained by random mutagenisation; its sequence was not characterised. In the comparison of different feedback-resistant strains obtained, AHAS activity of the strain carrying the ilvN CH allele was completely released from feedback inhibition in the presence of 10 mM l-valine, l-leucine and l-isoleucine. The plasmid pJC4 can be used as a shuttle vector in E. coli and in C. glutamicum as well as for the overexpression of genes in C. glutamicum (see [17] for an overview).
The strains were cultivated as described elsewhere [11]. A first preculture was inoculated from a cryoculture and grown in 500-ml shake flasks without baffles containing 50 ml LB medium [18] for 10 h. One millilitre was transferred to a second preculture with 50 ml CGXII minimal medium containing 225 mM (40 g/l) glucose and 100 mM (6 g/l) acetate in order to supplement the inactivation of the PDHC. After 14 h of growth, the second preculture was used to inoculate a bioreactor (Sixfors, Infors) containing 1.5 l CGXII medium, again with 225 mM glucose and 100 mM acetate. Cultivations were performed at 30°C at controlled pH of 7.2 and at a dissolved oxygen concentration of at least 30% maintained by controlling the stirrer velocity. The media contained 50 mg/l kanamycin in order to prevent the loss of the plasmids.
An additional acetate feed of 10 mM/h glacial acetic acid was added after 8 h to increase cell density for metabolome analysis.
Construction of the strain C. glutamicum ΔaceE pJC4ilvCDE
The plasmid pJC4ilvCDE was cloned as follows: The ilvBNC region was taken from C. glutamicum ATCC 13032 and amplified with primers BNCf2: ATACTAGCTAGCTAGGTACGCCTAAAATCATGAG and BNC-rev TATCCCTAGGTTCCTATGCGGCGGTTAAGC by polymerase chain reaction (PCR) with Phusion® Hot Start DNA polymerase (Biozym Scientific GmbH, D-31840 Hess, Oldendorf). The so-generated blunt-end PCR product was directly cloned into pCRbluntII (Invitrogen GmbH, Karlsruhe, Germany). The corresponding PCR product (4,130 bp) was cloned into pCRbluntII. The ilvD gene was taken from pECM3ilvBNCD [12] as 2,952-bp XbaI fragment and cloned into the NheI-cut pCRbluntII_ilvBNC, in divergent orientation to the ilvB gene, resulting in pCRbluntII_ilvBNCD. A 2.8-kb ilvBN fragment was then eliminated from pCRbluntII_ilvBNCD by cutting with BglII and Eco47III, treating the 7.76-kb linear pCRbluntII_ilvCD fragment with Klenow fragment and circularising it by religation, yielding pCRbluntII_ilvCD. A 4.3-kb AvrII/XbaI ilvCD fragment was then taken from pCRbluntII_ilvCD and cloned into XbaI-cut pJC4 [19]. Two orientations of ilvCD were possible. The ilvCD cluster was cloned such that the ilvC gene was in the same direction as the kanamycin-resistance gene of pJC4 vector. The ilvE gene was amplified with primers ilvE-XbaI_fw GCT CTA GAG CCA AGC CTA GCC ATT CCT CAA and ilvE-XbaI_rev GCT CTA GAG CCA GCC ACT GCA TTC TCC TTA to give a 1,384-bp fragment. After XbaI treatment, the ilvE fragment was cloned into pJC4_ilvCD that had been cut with NheI. Of two possible orientations of ilvE, we chose the one downstream of and in opposite direction to ilvD.
Analysis of biomass, substrate consumption and product formation
Biomass concentration was measured by optical density (OD) at λ = 546 nm (UV-160 spectrometer, Shimadzu). A factor of 0.332 was determined experimentally for the calculation of cell dry weight (CDW) from OD. Glucose was analysed using AccuChek sensor [20]. High-performance liquid chromatography (HPLC) analysis was used to analyse the concentrations of amino and organic acids [20, 21]. Three independent fermentations were performed to determine l-valine and by-product formation (two fermentations for the strain C. glutamicum ΔaceE).
Metabolome analysis
One technical and six analytical replicates were investigated in the metabolome analysis of PDHC-deficient strains harbouring different plasmids (C. glutamicum ΔaceE pJC4ilvBNCD, one analytical replicate). Samples were taken 4 h after the acetate feed had finished. Hence, a factor of 1.0 ml/g CDW was assumed as the cell volume for the calculation of intracellular concentrations [22]. Cytoplasmic metabolite concentrations were determined after inactivation of the metabolism by methanolic quenching [23]. Cell suspension (5 ml) was immediately quenched by transferring it into 15 ml methanol (60% at –50°C). The samples were centrifuged for 5 min at 10,000g at –20°C and the resulting pellet was resuspended in 1 ml methanol (−80°C) and 1 ml TE buffer [10 mM Tris, 1 mM ethylenediamine tetraacetic acid (EDTA); pH 7.0; 4°C]. After cell disruption by adding 2 ml chloroform and shaking the samples for 4 h at -20°C, the samples were analysed by liquid-chromatography tandem mass spectroscopy (LC-MS/MS) measurements using the modified method of Luo et al. [24]. The method was modified by the addition of the optimised MS/MS transitions of the intermediates of l-valine biosynthesis, i.e. acetolactate, DHIV and KIV. The quantification standard of acetolactate was synthesised directly before the measurement as described by Leyval et al. [4] due to the low stability of this intermediate. For the same reasons, samples were analysed directly after sampling to enable the acetolactate to be quantified.
The intracellular concentrations of the adenylate nucleotides adenosine monophosphate (AMP), adenosine diphosphate (ADP) and adenosine triphosphate (ATP) as determined by LC-MS/MS were used to calculate the energy charge. The energy charge (EC) was calculated according to Chapman et al. [25]: EC = [(ATP) + 1/2 (ADP)]/[(ATP) + (ADP) + (AMP)].
Results
Comparison of growth rate of l-valine production strains containing different plasmids
In order to gain a better understanding of l-valine biosynthesis and to identify an optimal plasmid for l-valine hyperproduction with reduced by-product formation, five PDHC-deficient strains overexpressing different combinations of l-valine biosynthesis genes were cultivated. Extracellular metabolite formation and intracellular concentration of l-valine intermediates were analysed. During the fermentations, no significant differences were observed on comparing the adenylate energy charges of these strains.
The energy charge was determined to be at least 0.80 for all strains, which is presumed to indicate growing cells [25]. It may be assumed that the leakage effect is different for different metabolites. However, the finding of an appropriate high energy charge in the metabolome analysis indicates reliable intracellular metabolite concentrations, since at least the sampling procedure was performed fast enough to prevent further conversion of metabolites. A low energy charge would give evidence that a remarkable part of ATP had been consumed during sample preparation to form ADP or AMP [26]. Hence, the metabolome data shown can be assumed valid for a better understanding of l-valine biosynthesis.
The growth rate of the wild-type C. glutamicum ATCC 13032 was determined as 0.44 1/h [11]. The growth rate of the PDHC-deficient strain was in the same range (0.46 1/h). Hence, the deletion of the aceE gene is shown to have no effects on the maximum growth rate, if acetate is sufficiently supplemented.
After the introduction of any plasmid, the additional burden from overexpression of l-valine biosynthesis genes led to a reduction of growth to μ max values around 0.39 1/h. Thus, the burden of different plasmids on the cell seems to be comparable and is not affected by the particular overexpressed genes. The feedback-resistant strain (ilvN CH) grew more slowly (μ max = 0.27 1/h), which was observed for several feedback-resistant l-valine production strains analysed in our laboratory (unpublished results).
l-Valine and extracellular by-product formation of different production strains
Considerable changes in extracellular metabolite concentrations (Fig. 2) resulting in different yields were observed as a consequence of the overexpression of different genes from l-valine biosynthesis (Table 1). Whereas no considerable formation of l-valine or any by-products were observed during growth phase, growth stopped with the depletion of acetate. Products and by-products were formed during the production phase under non-growing conditions with glucose as the only carbon source. With regard to differences in substrate consumption, especially for the strain harbouring the plasmid pJC4ilvBNCD, extracellular product and by-product concentrations are discussed as yield related to glucose consumed in millimol of product per mol glucose during production phase (mM/M).
For the wild-type C. glutamicum ATCC 13032, neither a remarkable formation of l-valine nor of any by-products has been reported [3, 11]. By contrast, the deletion of the aceE gene led to increased formation of pyruvate (269 mM/M) and l-alanine (240 mM/M). As much as 118 mM/M KIV and 115 mM/M l-valine were formed without modifications in the l-valine pathway. However, high by-product formation and the low flux to the desired product l-valine are insufficient, and carbon flux must be channelled more efficiently.
This was realised in the strain C. glutamicum ΔaceE pJC4ilvBNC by overexpression of the genes encoding the first two enzymes of l-valine biosynthesis, i.e. AHAS and AHAIR. As intended and expected the formation of the by-products was reduced. Pyruvate formation was almost abolished and l-alanine was reduced by 75% (60 mM/M). Compared with the PDHC-deficient strain without any plasmid, l-valine production was raised to 272 mM/M (corresponding to a concentration of 48.6 mM l-valine). Moreover, the formation of KIV also increased (394 mM/M). Hence, overexpression of the first part of the l-valine pathway is not sufficient to construct an efficient production strain.
Increased formation of pyruvate, but only slight reduction in l-alanine formation and even decreased formation of l-valine (22.4 mM l-valine instead of 48.6 mM), was observed after additional overexpression of the ilvD gene encoding the DHAD (plasmid pJC4ilvBNCD). In contrast, the accumulation of KIV increased to 580 mM/M. Hence, a limitation of carbon flux by transaminase B encoded by ilvE can be assumed in the observed genetic background. The strain harbouring the plasmid pJC4ilvBNCD showed strongly decreased glucose uptake, which resulted in reduced absolute concentrations of extracellular metabolites (Fig. 2).
Thus, the ilvE gene encoding for transaminase B seemed to be an important target to develop the optimal l-valine producer. Use of the plasmid pJC4ilvCDE led to a drop of KIV (151 mM/M) and an increase of l-valine formation (352 mM/M, compared with 272 mM/M using the plasmid pJC4ilvBNCD). However, 66 mM/M pyruvate and 191 mM/M l-alanine were further relevant by-products. Hence, overexpression of ilvBN is indispensable, and the first and the last enzymes of the l-valine pathway have to be overexpressed.
Formation of the l-valine precursor pyruvate was eliminated by overexpression of the genes encoding AHAS, AHAIR and transaminase B in the strain C. glutamicum ΔaceE pJC4ilvBNCE. Surprisingly, 42 mM/M l-alanine was still formed. Overexpression of ilvE prevented the accumulation of KIV, and 524 mM/M l-valine was formed, corresponding to absolute l-valine concentration of 104.3 mM.
An alternative to overexpression of the ilvBN genes encoding the AHAS was analysed by using the plasmid pJC4ilvCDE and a feedback-resistant allele of the AHAS gene in the bacterial chromosome. Compared with the strain harbouring the plasmid pJC4ilvBNCE, the l-valine yield increased only slightly to 536 mM with a feedback-resistant AHAS harbouring the plasmid pJC4ilvCDE. In contrast, the absolute l-valine concentration was slightly reduced from 104.3 to 98.7 mM. The comparison with the strain containing the plasmid pJC4ilvBNCE showed the minor function of DHAD (ilvD). However, 189 mM/M l-alanine was formed by the feedback-resistant strain. Hence, feedback resistance is not sufficient for effective redirection of pyruvate towards l-valine formation; overexpression of the ilvBN genes is necessary.
Intracellular analysis of l-valine intermediates in different PDHC-deficient strains
In parallel with the conventional analysis of the fermentation and extracellular metabolites, the concentration of l-valine intermediates was determined intracellularly (Table 2). The given cytoplasmic concentrations are based on the extraction of the cell pellet after quenching. Since the metabolites are also found extracellularly, the quenching supernatant was not taken into account to avoid the influence of extracellular metabolites.
Deletion of the aceE gene led to cytoplasmic accumulation of pyruvate (5.93 mM) and acetolactate (4.87 mM). As shown previously by Blombach et al. [3], inactivation of PDHC results in a high pyruvate pool. Pyruvate concentration was reduced by approximately 90% by overexpressing the genes coding for the first two enzymes of the l-valine pathway (plasmid pJC4ilvBNC). A value of 4.87 mM acetolactate was found in the PDHC-deficient strain without plasmid, which was reduced by 43% by ilvBNC overexpression. Since the DHIV (from 0.03 to 4.91 mM) and the KIV pool (from 1.24 to 6.76 mM) were dramatically increased after ilvBNC overexpression, amplification of the first part of the pathway alone is insufficient for efficient l-valine formation.
As expected, the strain containing the plasmid pJC4ilvBNCD for additional overexpression of DHAD encoded by ilvD showed reduced intracellular DHIV concentrations (below detection limit), but also further accumulation of intracellular KIV up to 25.95 mM.
As already concluded from extracellular measurements, the ilvE gene encoding for transaminase B seems to be an important target for an optimal l-valine producer. After overexpression, cytoplasmic KIV was reduced to 1.72 mM using the plasmid pJC4ilvCDE and to 1.75 mM with an additional feedback-resistant ilvN allele. Comparable amounts of acetolactate and DHIV were detected for both strains, indicating the minor influence of ilvD. The feedback-resistant AHAS reduced intracellular pyruvate from 11.07 mM to 0.84 mM.
The l-valine production strain C. glutamicum ΔaceE pJC4ilvBNCE displayed low cytoplasmic concentrations, with 0.79 mM pyruvate and 2.07 mM KIV. Surprisingly, the accumulation of 10.51 mM acetolactate and 13.67 mM DHIV exceeded that for the other strains.
Discussion
PDHC-deficient strains overexpressing different genes of l-valine biosynthesis were analysed in order to develop an efficient fermentative l-valine production process with reduced by-product formation. An appropriate plasmid was to be identified by comparing the concentrations of cytoplasmic intermediates and extracellular by-products.
After precursor accumulation achieved by PDHC inactivation [3], pyruvate is now to be channelled towards l-valine. It can be concluded from the present results that ilvBN overexpression is essential to achieve sufficient carbon flux towards the desired product. Using the plasmids overexpressing these genes encoding acetolactate synthase (pJC4ilvBNC, pJC4ilvBNCD and pJC4ilvBNCE), strong reduction of cytoplasmic pyruvate and extracellular l-alanine was observed. The need for ilvBN overexpression is consistent with earlier approaches [12, 15]; it can be explained by competing enzymes using pyruvate, especially in l-valine-overproducing strains. After PDHC inactivation and subsequent increased intracellular pyruvate concentration, particularly l-alanine- and l-valine-forming enzymes compete for pyruvate. The K m for pyruvate of l-alanine-forming l-valine-pyruvate aminotransferase (0.41 mM) is comparatively low compared with the K m of AHAS (8.3 mM) [4, 27]. Therefore, l-alanine formation is favoured over l-valine formation. Thus, overexpression of ilvBN reduces pyruvate availability for l-alanine formation and therefore results in l-alanine reduction. The benefit of overexpressing ilvBN is that it results in an intracellular pyruvate concentration decreased by at least 85%. However, an exception is found with 1.48 mM cytoplasmic pyruvate in the strain using the plasmid pJC4ilvBNCD. This can be explained by the activity of the l-valine-pyruvate aminotransferase. In the presence of high amounts of KIV without ilvE overexpression, the reaction equilibrium of this transaminase is shifted towards pyruvate and l-valine. Hence, also less l-alanine can be detected.
The beneficial effect of ilvE overexpression avoiding the pyruvate-forming reaction of l-valine-pyruvate aminotransferase was only found in PDHC-deficient strains. A KIV yield on glucose of 580 mM/M and less l-valine were formed without ilvE overexpression, which was avoided by the amplification of ilvE. Overexpression of ilvE encoding transaminase B was hitherto assumed to be dispensable. Radmacher et al. [12] showed reduced l-valine formation with transaminase B overexpression when comparing the plasmids pJC1ilvBNCD and pJC1ilvBNCE. Hence, it can be concluded that overexpression of this gene is beneficial for l-valine formation, at least in the given genetic background, although it may not be necessary in l-valine production strains with active PDHC. Inactivation of PDHC impedes the carbon flux into the tricarboxylic acid cycle (TCA) and thus prevents the isocitrate dehydrogenase reaction. This reaction is one of four NADPH-forming reactions in C. glutamicum [13]. Therefore, NADPH limitation can be assumed in PDHC-deficient l-valine production strains since the pentose phosphate pathway is the only NADPH source left after PDHC inactivation, and two moles of NADPH are consumed on l-valine formation [13]. The limitation in NADPH may result in KIV accumulation. TCA activity and hence higher NADPH availability can be assumed for the pantothenate and isoleucine auxotrophic l-valine producers analysed by Radmacher et al. [12].
Combining overexpression of the ilvBNCE genes, intracellular concentrations of acetolactate and DHIV were comparatively high. Since the AHAIR encoded by ilvC is also NADPH-consuming, efficient l-valine formation due to ilvBNCE overexpression may result in NADPH limitation, resulting in intracellular acetolactate accumulation.
Compared with the wild-type allele and the plasmid pJC4ilvBNCE, a feedback-resistant AHAS in combination with the overexpression of the genes ilvCDE did not increase l-valine formation. It can be concluded that AHAS overexpression can overcome feedback regulation. However, the amplification of genes encoding a feedback-resistant AHAS should be analysed.
Overexpression of all genes encoding for the l-valine forming pathway may be another target for further research. However, the ilvD gene encoding DHAD may be omitted, as shown by the comparatively low intra- and extracellular KIV concentrations after ilvE overexpression. This conclusion was also drawn by Magnus et al. [28] when determining the flux control coefficients for the l-valine pathway. The main flux control for l-valine formation was through AHAS, transamination and l-valine export. A tenfold decrease in DHAD activity is calculated to reduce l-valine yield by just 4.3%.
It can be concluded from the results obtained that the ideal combination of overexpressed genes in a certain pathway depends on the relative genetic background. Using different genetic modifications for l-valine hyperproduction results in the need for overexpression of different groups of genes. Hence, the composition of an optimal plasmid has to be revised after additional genetic modifications. Cofactor availability affects the pathway and must be involved in strain design. Metabolome analysis is a good and comparatively fast method for plasmid optimisation, if more information has to be obtained than is available from conventional cultivations analysing only the extracellular metabolite pattern. Calculation of flux control coefficients may be an alternative; however, it is much more time consuming.
References
Gavrilescu M, Chisti Y (2005) Biotechnology—a sustainable alternative for chemical industry. Biotechnol Adv 23:471–499
Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69:1–8
Blombach B, Schreiner ME, Holatko J, Bartek T, Oldiges M, Eikmanns BJ (2007) L–Valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl Environ Microbiol 73:2079–2084
Leyval D, Uy D, Delaunay S, Goergen JL, Engasser JM (2003) Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J Biotechnol 104:241–252
Kennerknecht N, Sahm H, Yen MR, Patek M, Saier MH, Eggeling L (2002) Export of L–isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956
Ebbighausen H, Weil B, Krämer R (1989) Transport of branched-chain amino-acids in corynebacterium-glutamicum. Arch Microbiol 151:238–244
Sugisaki Z (1959) Studies on L–valine fermentation. Part 1- Production of L–valine by Aerobacter Bacteria. J Gen Appl Microbiol 5:138–149
Chatterjee M, Chatterjee SP (1982) Valine production from hydrocarbon by Micrococcus-Varians. Folia Microbiol 27:228–236
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Park JH, Lee KH, Kim TY, Lee SY (2007) Metabolic engineering of Escherichia coli for the production of L–valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA 104:7797–7802
Bartek T, Makus P, Klein B, Lang S, Oldiges M (2008) Influence of L–isoleucine and pantothenate auxotrophy for L–valine formation in Corynebacterium glutamicum revisited by metabolome analyses. Bioprocess Biosyst Eng 31:217–225
Radmacher E, Vaitsikova A, Burger U, Krumbach K, Sahm H, Eggeling L (2002) Linking central metabolism with increased pathway flux: L–valine accumulation by Corynebacterium glutamicum. Appl Environ Microbiol 68:2246–2250
Bartek T, Blombach B, Zönnchen E, Makus P, Lang S, Eikmanns BJ, Oldiges M. Importance of NADPH supply for improved L–valine formation in Corynebacterium glutamicum. Accepted for publication in Biotechnol Prog
Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ (2008) Corynebacterium glutamicum tailored for high-yield L–valine production. Appl Microbiol Biotechnol 79:471–479
Elisakova V, Patek M, Holatko J, Nesvera JN, Leyval D, Goergen JL, Delaunay S (2005) Feedback-resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum. Appl Environ Microbiol 71:207–213
Schreiner ME, Fiur D, Holatkoj J, Patek M, Eikmanns BJ (2005) E1 enzyme of the pyruvate dehydrogenase complex in Corynebactetium glutamicum: Molecular analysis of the gene and phylogenetic aspects. J Bacteriol 187:6005–6018
Eggeling L, Reyes O (2005) Experiments. In: Bott M, Eggeling L (eds) Handbook of Corynebacterium glutamicum. Taylor & Francis, Boca Raton, London, pp 535–566
Sambrook J, Russel DW (2001) Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Cremer J, Eggeling L, Sahm H (1990) Cloning the dapa dapb cluster of the lysine-secreting bacterium corynebacterium-glutamicum. Mol Gen Genet 220:478–480
Brik-Ternbach M, Bollman C, Wandrey C, Takors R (2005) Application of model discriminating experimental design for modeling and development of a fermentative fed-batch L–valine production process. Biotechnol Bioeng 91:356–368
Zelic B, Gostovic S, Vuorilehto K, Vasic-Racki B, Takors R (2004) Process strategies to enhance pyruvate production with recombinant Escherichia coli: from repetitive fed-batch to in situ product recovery with fully integrated electrodialysis. Biotechnol Bioeng 85:638–646
Rönsch H, Krämer R, Morbach S (2003) Impact of osmotic stress on volume regulation, cytoplasmic solute composition and lysine production in Corynebacterium glutamicum MH20–22B. J Biotechnol 104:87–97
de Koning W, van Dam K (1992) A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal Biochem 204:118–123
Luo B, Groenke K, Takors R, Wandrey C, Oldiges M (2007) Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A1147:153–164
Chapman AG, Fall L, Atkinson DE (1971) Adenylate energy charge in Escherichia-Coli during growth and starvation. J Bacteriol 108:1072–1086
Bolten CJ, Kiefer P, Letisse F, Portais J-C, Wittmann C (2007) Sampling for metabolome analysis of microorganisms. Anal Chem 79:3843–3849
Ambartsumyan AA, Bezirdzhyan KO (1994) Catalytic properties of valine—pyruvate aminotransferase from brevibacterium-flavum. Biochemistry-Moscow 59:1027–1032
Magnus JB, Oldiges M, Takors R (2009) The identification of enzyme targets for the optimization of a valine producing Corynebacterium glutamicum strain using a kinetic model. Biotechnol Prog 25:754–762
Acknowledgments
This work was financially supported by the Fachagentur Nachwachsende Rohstoffe (Agency for Renewable Resources) of the BMVEL—German Federal Ministry of Food, Agriculture and Consumer Protection (grant 04NR003/22000304) and by Evonik Degussa GmbH. The authors would like to thank Andreas Karau (Evonik Degussa GmbH), Bastian Blombach and Bernard J. Eikmanns (Ulm University) for fruitful cooperation and the valuable discussion of results.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bartek, T., Zönnchen, E., Klein, B. et al. Analysing overexpression of l-valine biosynthesis genes in pyruvate-dehydrogenase-deficient Corynebacterium glutamicum . J Ind Microbiol Biotechnol 37, 263–270 (2010). https://doi.org/10.1007/s10295-009-0669-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0669-x