Abstract
To gain insight into the impact of 2,4,6-trinitrotoluene (TNT) on soil microbial communities, we characterized the bacterial community of several TNT-contaminated soils from two sites with different histories of contamination and concentrations of TNT. The amount of extracted DNA, the total cell counts and the number of CFU were lower in the TNT-contaminated soils. Analysis of soil bacterial diversity by DGGE showed a predominance of Pseudomonadaceae and Xanthomonadaceae in the TNT-contaminated soils, as well as the presence of Caulobacteraceae. CFU from TNT-contaminated soils were identified as Pseudomonadaceae, and, to a lesser extent, Caulobacteraceae. Finally, a pristine soil was spiked with different concentrations of TNT and the soil microcosms were incubated for 4 months. The amount of extracted DNA decreased in the microcosms with a high TNT concentration [1.4 and 28.5 g TNT/kg (dry wt) of soil] over the incubation period. After 7 days of incubation of these soil microcosms, there was already a clear shift of their original flora towards a community dominated by Pseudomonadaceae, Xanthomonadaceae, Comamonadaceae and Caulobacteraceae. These results indicate that TNT affects soil bacterial diversity by selecting a narrow range of bacterial species that belong mostly to Pseudomonadaceae and Xanthomonadaceae.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
2,4,6-Trinitrotoluene (TNT) is an environmental pollutant found at many sites associated primarily with military and, occasionally, with civil activities. It is known to be toxic against various eukaryotic organisms like fungi, algae or worms. The toxicity of TNT is frequently caused by the enzymatic single-electron reduction of its nitro groups, which leads to the formation of reactive oxygen species in living cells [41 and references therein]. However, few reports describe the effect of TNT on soil bacterial diversity. Wilke et al. [47] found only minor differences between TNT-contaminated and uncontaminated soils by phospholipid fatty acid composition analysis. Likewise, in the study of Wikström et al. [46], random amplified polymorphic DNA analysis did not reveal any significant difference in the composition of lake microbial communities due to the presence of TNT. Conversely, Fuller and Manning [15] noticed a predominance of Gram-negative over Gram-positive bacteria in TNT-contaminated soils. Using respirometric activities, Gong et al. [17] observed a decrease in the specific growth rate and an increase in organic matter content in soils with a high TNT concentration. A decrease in denitrification and dehydrogenase activities and a low microbial biomass carbon were also reported in TNT-contaminated soils [30,38]. All in all, these results suggest that TNT affects to a variable extent soil bacterial composition and activity. However, the bacterial composition at the group and species level was only partially resolved in the aforementioned studies, because the techniques used were not specific enough to detail the bacterial community composition of TNT-contaminated environments.
16S rRNA and its gene are powerful markers to identify and classify bacteria. Various molecular methods based on the 16S rRNA gene have been developed to characterize the bacterial composition of environmental samples. Among those, the separation of PCR-amplified 16S rRNA genes using denaturing gradient gel electrophoresis (DGGE) is a widely used methodology that provides a direct fingerprint of environmental samples [32]. In addition, bands can be excised from the gel and sequenced to identify the dominant species in the community analyzed. Consequently, DGGE has been used by several research groups to assess soil microbial diversity and shifts in communities due to xenobiotic contamination, e.g. [5, 6, 11, 13, 28].
Over the past two decades, the use of such cultivation-independent methods has rapidly expanded over traditional cultivation-dependent approaches. This has been primarily due to the limitations of the cultivation approach, as most of the bacteria remain uncultured using traditional laboratory techniques. The proportion of culturable bacteria has been estimated at less than 1% of soil bacteria [43]. However, recent developments in cultivation techniques have allowed to increase the proportion of culturable cell counts and to isolate previously unculturable microorganisms [14, 20, 26] and references therein, and [42]. In some polluted environments, adapted cultivation-dependent methods even proved to be superior to cultivation-independent methods, because they reflected the effect of the pollutant on the physiological status of bacteria [7].
This study was conducted to assess the impact of TNT contamination on soil bacterial communities using both cultivation-independent and -dependent methods. Changes in soil bacterial diversity due to TNT contamination were investigated at two sites (Bourges, France and El Gordo, Spain) with different histories of TNT contamination, different TNT concentrations as well as different physico-chemical characteristics. DGGE analyses were carried out to provide a fingerprint of bacterial communities in TNT-contaminated soils compared to non-contaminated ones, and to identify the major species present in TNT-contaminated soils. In addition, bacteria from these soils were cultivated on a solid dilute medium in order to determine whether TNT contamination had patterned their cultivable community as well. Finally, to test if the conclusions drawn from these experiments were valid for other soils as well, a pristine soil sample from our campus was spiked with different concentrations of TNT and changes in its bacterial community composition were monitored over time.
Methods
Soil characteristics
Two series of soils were investigated. The first one (samples REF, KX1, KL1, KL2, KF1, KF2, KF4, KF5, KF6, KF6b, KF6c, F1, F2, and F3) was collected at a site in Bourges, France that had been used for TNT destruction over the past 20 years. Samples were taken at different locations at the same site with a distance between them of approximately 10 m. Uncontaminated (i.e., TNT was not detected by HPLC with a detection limit of 1 mg TNT/kg soil) soil samples were collected in an area shielded from heavy TNT contamination by the presence of a protecting wall. A second series of soil samples was provided by Fabricaciones Extremeñas S.L., a subsidiary company of Union Española de Explosivos (Madrid), from a site located at El Gordo, Cáceres, Spain. It was artificially contaminated with TNT for 7 months and separated into different plots. Two TNT-contaminated soil samples were taken from a contaminated plot (samples UEE and IN). Another TNT-contaminated plot was planted with corn seeds coated with Pseudomonas putida sp. JLR11, a strain able to use TNT as N-source and terminal electron acceptor [10], and samples were taken 15 (sample J15) and 29 (sample J29) days after planting. A third TNT-contaminated plot was planted with non-coated corn seeds and samples were collected 15 (sample C15) and 29 (sample C29) days after planting. One sample (sample OUT) was collected at the same site from a non-contaminated plot. For the two series, samples were collected from the top 10-cm layer of soil, except samples KF6b and KF6c, which were taken at a depth of 30 cm. Finally, in order to evaluate the effect of TNT on a pristine soil, a soil from Louvain-la-Neuve (Belgium) was artificially contaminated with increasing concentrations of TNT.
All samples were homogenized, sieved (2-mm mesh) to eliminate plant roots and stones, and stored at 4°C in sterile containers. The concentration of TNT (and its metabolites) in the soil samples was determined in triplicates as previously described [11]. Soil pH was measured as follows: 1 g of soil was mixed with 5 g of demineralized water, blended and allowed to settle for 20 min three times in a row. After the third settling, the pH of the supernatant was measured. Soil texture was determined using the pipette method after removal of carbonate and organic matter with HCl and H2O2, respectively, and dispersion with hexametaphosphate [24]. The soil carbon content was measured in triplicate by chemical oxidation with concentrated H2SO4 and K2Cr2O7 [1].
Four soils collected at the Bourges site (REF, KF4, F1, and F2) were uncontaminated and ten (KX1, KL1, KL2, KF1, KF2, KF5, KF6, KF6b, KF6c and F3) were contaminated with high concentrations of TNT [from 3.59 to 36.2 g TNT/kg (dry wt) of soil]. In the TNT-contaminated soils, the concentrations of aminodinitrotoluenes (4-A-2,6-DNT or 2-A-4,6-DNT), 2,4,6-trinitrobenzaldehyde (TNBA), and 2,4-dinitrotoluene (2,4-DNT) were very low compared to the concentration of TNT, as they ranged from <0.001 [detection limit of HPLC analysis to a maximum of 0.065 g/kg (dry wt) of soil]. All samples had a pH above 7.6. These soils were classified as sandy (clay) loams or (loamy) sands, except for sample KF4, which was mostly silty. The percentage of carbon varied significantly between the samples, with values ranging between 0.7 and 14.5% C. On the contrary, the TNT-contaminated soils collected at the site of El Gordo contained relatively low concentrations of TNT [from 0.01 to 1.57 g TNT/kg (dry wt) of soil]. They were slightly acidic (pH between 5.2 and 6.5) loamy sands. Their organic matter content averaged 0.8% (P. van Dillewijn, personal communication). The soil from Louvain-la-Neuve was a silt loam with a pH of 7.4. Soil characteristics are detailed in Table 1.
Cultivation experiments
One gram of soil REF or KX1 was added to 100 ml of sterile dH2O (100-fold dilution) and stirred for 2 h at 300 rpm. After settling for 1 h, the supernatant was serially diluted tenfold in sterile dH2O (1,000–100,000-fold final dilutions). Aliquots (150 μl) of each dilution were spread in three to five replicates onto agar plates supplemented with dilute nutrient broth [containing per liter: 15 g agar (Sigma, Saint Louis, MO) and 0.065 g nutrient broth (Oxoïd, Hampshire, UK) (i.e., 200 times less than the manufacturer’s instructions)] with a sterile glass-spreading rod. Plates were incubated for up to 3 months to allow the emergence of slow-growing bacteria. The plates were incubated in the dark at room temperature. The number of colony forming units (CFU) on each medium was counted 2, 7, 8, 14, 22, 35, 49, 63, 77, and 91 days after inoculation. For each time point, plates carrying between 50 and 250 colonies were used to calculate the number of colonies per g of soil. After 14, 35, 63 and 91 days, plates corresponding to the 1,000-fold dilution were washed with 1.5 ml of sterile dH2O. The washing solution was centrifuged (10,000g for 5 min) and the pellets were used for DNA extraction and plate-wash PCR [42].
Spiking of a pristine soil with TNT
A pristine soil was sampled at the University campus of Louvain-la-Neuve, Belgium. It was sieved on a 2-mm mesh and divided into four aliquots: a control sample (no TNT), and three samples spiked with increasing concentrations of TNT: 140 mg, 1.4 g, and 28.5 g TNT/kg (dry wt) of soil. TNT was crushed with a mortar until a very thin powder was obtained, that was very carefully mixed with the soil material. Seven hundred and fifty g of each of these four samples was poured into 1–l beakers covered with aluminium foil and incubated in the dark at room temperature over 4 months. Each of them constituted an individual microcosm for bacterial community observations. Soil samples had an initial water content of 32.5% (measured in triplicates by drying 3 g of wet soil at 105°C up to constant weight). The water content was kept constant in the microcosms by monitoring every 15 days the percentage of humidity and readjusting it with sterile dH2O if necessary. For each microcosm, 1 g of soil was collected for DNA extraction a few minutes after the addition of TNT (t = 0), and 7, 14, 61, and 121 days after the addition of TNT.
DNA extraction
DNA was extracted from 1 g of soil using the Ultra-Clean Soil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer’s instructions. The DNA of plate washed CFU was extracted from the pellets using the same kit, except that the bead-beating solution was supplemented with a 0.22-μm-filtered solution of lysozyme (5 mg/ml final concentration) and the pellet was incubated in this solution overnight at 37°C. Moreover, the Inhibitor Removal Solution (IRS), provided by the manufacturer to precipitate humic acids, was not used. The quantity and quality of the DNA extracted was evaluated by gel electrophoresis on 1% agarose gels stained with ethidium bromide. About 5–10 μl of eluted DNA were loaded on the gels. DNA was quantified using the free software Image J (available at http://rsb.info.nih.gov/ij/).
Total cell counts
TNT-contaminated and uncontaminated soil samples were fixed, dispersed by ultrasonication and stained with 4′,6-diaminido-2-phenylindole (DAPI) following the protocol of Janssen et al. [20]. The microscope slides were examined with a Leica microscope DMRA2 (Leica Microsystems GmbH, Wetzlar, Germany) under UV illumination (excitation filter BP340-380, dichromatic mirror: 400, suppression filter, LP425). Blue-fluorescing cells were counted in forty fields for each sample and expressed as cells/g (dry wt) of soil.
PCR amplification
16S rRNA genes were amplified using primers BACT63F (5′-CAGGCCTAACACATGCAAGTC-3′, forward) [29] and UNIV518R (5′-ATTACCGCGGCTGCTGG-3′, reverse) [32] with a 40 bp GC-clamp attached on the forward primer. BACT63F complements a conserved region in the domain Bacteria whereas UNIV518R complements a universally conserved region. The PCR mix (50 μl) contained 25 μl of Red’y’star Mix (Eurogentec, Seraing, Belgium), 0.25 μM of each primer and ∼2–10 ng of target DNA. For some soil samples with a low DNA extraction yield, DNA quantification was not possible using gel electrophoresis. Therefore, different volumes of extracted DNA were tested, and in all cases 1–2 μl gave the best PCR signals. PCR was performed using a Biometra TGradient cycler (Biometra, Göttingen, Germany) under the following conditions: 95°C for 5 min, followed by 30 cycles (95°C for 1 min, 55°C for 1 min, 72°C for 1 min), with a final extension at 72°C for 10 min. PCR products were visualized by gel electrophoresis on 1.5% agarose gels stained with ethidium bromide and quantified with the Image J software.
DGGE analysis and sequencing of DGGE fragments
DGGE analysis was carried out with the DCode universal mutation detection system (Bio-Rad, Hercules, CA). PCR products were loaded onto 6% (wt/vol) polyacrylamide (37.5:1 acrylamide:bisacrylamide) gels with a linear denaturing gradient from 35 to 55% (where 100% contains 7 M urea and 40% (vol/vol) deionized formamide) in 0.5 × TAE buffer (20 mM Tris base, 10 mM acetate, 0.5 mM EDTA [pH 8]). Three hundred to 650 ng of PCR products were loaded onto the gel, except for samples KF6b and KF6c. For the latter, PCR amplification gave low yields, and the largest volume of PCR product was loaded onto the DGGE gel, which corresponded to 150 ng of PCR product. The electrophoresis was run at 60°C and 185 V for 5 h30 min. Gels were stained with ethidium bromide and photographed under a UV transilluminator.
Gel images were converted, normalized, and analyzed with the Bionumerics 4.6 software package (Applied Maths, Sint-Martens-Latem, Belgium) using the Pearson (product-moment) correlation coefficient and the unweighted pair group clustering method with arithmetic averages (UPGMA). Fingerprint analysis using the Pearson correlation coefficient, which calculates similarity indices of the densitometric curves of the DGGE fingerprints, is considered a more robust approach than band matching [18]. Different reference bands were used for the DGGE gels of the soils from Bourges and El Gordo and the pristine soil spiked with TNT. Therefore, they could not be compared in a single cluster analysis.
Bands of interest were excised from the gel and incubated overnight at 4°C in 35 μl of sterile dH2O. Next, 1–8 μl of this solution was used for a subsequent PCR amplification (40 μl volume) with 20 μl of ReadyMix (Sigma) and 0.25 μM of primers BACT63F and UNIV518R. The same amplification cycle was used as described above, except that 25 cycles were performed instead of 30. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Sequencing was carried out using an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) and the BigDye Terminator V1.1 cycle sequencing kit (Applied Biosystems). The 20-μl sequencing mixture contained 15–30 ng of purified PCR product and 0.5 μM of primer BACT63F or UNIV518R. This sequencing protocol gave unambiguous sequences for most DGGE bands. When ambiguous, the DGGE bands were cloned prior to sequencing using the pDrive vector (Qiagen).
Comparative sequence analysis and construction of the phylogenetic tree
Sequences corresponding to DGGE bands were aligned with about 43,000 homologous prokaryotic 16S rRNA gene sequences by using the automated aligning tool of the ARB program package (http://www.mikro.biologie.tu-muenchen.de, “ssu_jan04_corr_opt.arb” database) [27]. The topology of the groups, alpha-, beta-, gamma1- and gamma2-Proteobacteria in the tree containing all sequences >1,400 bp (“tree_1400_jan05”) was optimized using the global optimisation tool and appropriate filters. Then the partial sequences were inserted into the optimized tree using an ARB parsimony tool that did not affect the initial tree topology. The resulting tree was split into two subtrees and pruned for the clarity of the figures, and close reference organisms were retained.
Nucleotide sequence accession numbers
Sequences have been deposited in the GenBank database under accession numbers EF121875 to EF121949.
Results and discussion
DNA extraction yield, total cell counts and CFU counts in uncontaminated and TNT-contaminated soils
The amount of soil DNA extracted from the soils from Bourges and El Gordo was inversely related to their level of contamination. It was calculated by electrophoresis gel analysis that the uncontaminated soils from Bourges (REF, KF4, F1, and F2) and El Gordo (OUT) contained 1.8–3.3 μg of DNA/g (dry wt) of soil \( (\bar{X},2.48; \) SD, 0.58). DNA extracted from the moderately contaminated soils from El Gordo (UEE, C15, J15, C29, J29) ranged between 0.15 and 0.82 μg of DNA/g (dry wt) of soil \( (\bar{X},0.50; \) SD, 0.20). In some contaminated soils from Bourges (KL2, KL2, F3), it ranged between 0.86 and 0.97 μg of DNA/g (dry wt) of soil \( (\bar{X},0.92; \) SD, 0.06). Finally, the amount of DNA extracted from the most heavily contaminated soils from Bourges (KX1, KF1, KF2, KF5, KF6, KF6b, and KF6c) and El Gordo (IN) could not be quantified by gel analysis, because DNA was hardly visible or not visible at all on the agarose gels. Yet, this did not hamper 16S rRNA gene PCR-DGGE analysis of those soils (Figs. 1a, 2a). DNA quantification data suggested a lower bacterial biomass in TNT-contaminated soils, or an inhibitory effect of TNT on the extraction of DNA with the Mo Bio kit. The former hypothesis was confirmed by total cell counts in four uncontaminated soils (OUT, REF, KF4 and the pristine soil from Louvain-la-Neuve) and seven TNT-contaminated soils (KX1, KL2, KF5, KF6, UEE, IN, and J29). The number of DAPI-stained cells was significantly higher in uncontaminated soils \( [\bar{X}, \) 2.09 × 109 cells/g (dry wt) of soil; SD, 1.12 × 109, n = 4] than in TNT-contaminated ones \( [\bar{X}, \) 3.23 × 108 cells/g (dry wt) of soil; SD 1.85 × 108, n = 9] (one-tailed t-test, P = 0.0015). Moreover, the hypothesis that TNT affects the efficacy of DNA extraction was disproven by an experiment where a pristine soil was spiked with increasing amounts of TNT. At the beginning of the experiment (i.e. just after the addition of TNT), the amount of DNA in the four soil microcosms spiked with different concentrations of TNT was similar \( [\bar{X}, \) 1.55 μg of DNA/g (dry wt) of soil; SD 0.24] and remained relatively stable over a 61-day period. After 4 months of incubation, a reduction of 11 and 12% of the initial DNA content was measured in the control soil (no TNT added) and in the soil containing 140 mg TNT/kg soil, respectively. In the soils containing 1.4 and 28.5 g TNT/kg soil, the reduction amounted to 79% of the initial DNA content. These results indicate that the concentration of TNT and the duration of contamination affected soil bacterial biomass and therefore the amount of DNA extracted from those soils.
Genetic analysis of the bacterial community of soils collected in Bourges, France. a DGGE fingerprints of 16S rRNA gene amplicons derived from uncontaminated (REF to F2) and TNT-contaminated soils (F3 to KF6c). Numbers correspond to bands that were excised and sequenced. Their assignment to taxonomic groups is presented in Figs. 5 and 6. b Dendrogram of the DGGE fingerprints calculated by the Pearson correlation and the UPGMA method
Genetic analysis of the bacterial community of soils collected in El Gordo, Spain. a DGGE fingerprints of 16S rRNA gene amplicons derived from uncontaminated (OUT) and TNT-contaminated soils (UEE to J29). Numbers correspond to bands that were excised and sequenced. Their assignment to taxonomic groups is presented in Figs. 5 and 6. b Dendrogram of the DGGE fingerprints calculated by Pearson correlation and the UPGMA method
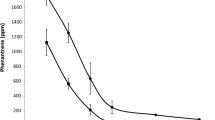
TNT influenced the number of soil culturable bacteria as well. Cultivation experiments were carried out with an uncontaminated (REF) and a TNT-contaminated soil (KX1), which had relatively similar pH, soil texture and carbon content values. Soil dilutions were plated on nutrient broth solidified with agar (“diluted NB”) and incubated for 13 weeks. Such experimental conditions had been reported to favor the cultivation of a larger proportion of soil microbes, including bacteria that were formely known as unculturable, like members of the division Actinobacteria, Acidobacteria and Verrucomicrobia [20]. A clear difference in the number of CFU was observed between the uncontaminated and the TNT-contaminated soil (Fig. 3a). The viable count profile (number of CFU as a function of incubation time) obtained with the uncontaminated soil REF was similar to profiles observed by Janssen et al. [20] with pasture soils, i.e., a gradual increase over a 91-day period of incubation. Conversely, for soil KX1 the number of CFU was stable after 21 days. After 91 days of incubation, the number of CFU reached 2.28 × 107 CFU/g (dry wt) of soil REF versus 1.15 × 107 CFU/g (dry wt) of soil KX1, i.e. 1 and 2% of total cell counts, respectively. This experiment was repeated and gave very similar results (data not shown). After 91 days of incubation, the late-appearing colonies had a smaller diameter than those appearing early on the plates, but the size of bacterial colonies was small in any case, as it ranged from ∼0.5 to ∼2 mm diameter. Plating dilutions of the uncontaminated soil KF4 versus the TNT-contaminated soil KF6 also showed a significantly higher number of CFU in the former after 91 days of incubation (4.44 × 107 CFU/g (dry wt) of soil KF4 versus 2.64 × 106 CFU/g (dry wt) of soil KF6), and the CFU curve reached a plateau after 21 days for the TNT-contaminated soil KF6 (data not shown).
Analysis of the cultivable microflora of one uncontaminated soil (REF) and one TNT-contaminated soil (KX1). a Evolution of the number of CFU per g of dry soil on agar plates supplemented with dilute nutrient broth as a function of incubation time. Closed and open squares are CFU from soil REF and KX1, respectively. Error bars represent the standard deviation of triplicates to quintuplicates of the same dilution. DGGE fingerprints of 16S rRNA gene amplicons derived from the cultivable bacterial microflora of soil REF (b) and soil KX1 (c) after 14, 35, 63 and 91 days of incubation. The first lane shows the DGGE fingerprint of 16S rRNA genes directly amplified from the two soils (as in Fig. 1a). Numbers correspond to bands that were excised and sequenced. Their assignment to taxonomic groups is presented in Figs. 5 and 6
Taken together, these results demonstrate the adverse effect of TNT on bacterial biomass in soils (expressed as the amount of extracted DNA, the total number of cells, or the number of CFU). They corroborate the observations by Fuller and Manning [15] and Meyers et al. [30] that the number of CFU developing on rich or moderately diluted agar media is low in soils severely contaminated with TNT. Likewise, in metal-contaminated soils, Ellis et al. [7] observed a lower number of cultivable bacteria in the samples with the greatest metal content.
Total and culturable bacterial diversity in TNT-contaminated soils
The bacterial diversity of the historically contaminated soils from Bourges and El Gordo was investigated by DGGE after PCR amplification of 16S rRNA gene fragments using primers BACT63F and UNIV518R. In a previous and more limited study, Eyers et al. [12] had analyzed the bacterial community of two soils from Bourges (the uncontaminated soil REF and the TNT-contaminated soil KX1) by hybridization of in vitro-transcribed 16S rRNA to a phylogenetic oligonucleotide microarray, and they had found a significant difference in hybridization patterns between these two soil samples. However, the origin of such a difference (e.g. presence of TNT, pH, soil texture, carbon content) had not been clearly determined. In the present study, the analysis of TNT-contaminated soils was extended to numerous soil samples from two contaminated sites as well as to soil microcosms artificially contaminated with TNT, in order to determine whether TNT systematically affected soil bacterial diversity.
The uncontaminated soils from Bourges (REF, KF4, F1, and F2) and El Gordo (OUT) showed a complex DGGE fingerprint consisting in a smear of bands, with none of them dominating the fingerprint, except two bands for soil F1 (Figs. 1a, 2a). Likewise, the soils microcosms with no TNT or 140 mg TNT/kg harboured complex DGGE fingerprints that did not change during the 121-day incubation period (Fig. 4a).
Genetic analysis of the bacterial community of a pristine soil spiked with TNT. a DGGE fingerprints of 16S rRNA gene amplicons derived from the pristine soil at t = 0 and 7, 14, 61 and 121 days after artificial contamination with different concentrations of TNT. Numbers correspond to bands that were excised and sequenced. Their assignment to taxonomic groups is presented in Figs. 5 and 6. b Dendrogram of the DGGE fingerprints calculated by Pearson correlation and the UPGMA method
Conversely, the DGGE fingerprints of the TNT-contaminated soils from Bourges (F3, KX1, KL1, KL2, KF1, KF2, KF5, KF6, KF6b, and KF6c) and El Gordo (UEE, IN, C15, J15, C29, J29) clearly revealed the presence of a few dominant bands (Figs. 1a, 2a). Duplicate samples of soils yielded similar DGGE patterns (data not shown). Cluster analysis revealed a similarity of 60% between eight (out of ten) TNT-contaminated soils from Bourges (Fig. 1b). The heavily contaminated soil KF6b had a low similarity (37%) with the other TNT-contaminated soils, but this was probably due to the very weak intensity of its DGGE fingerprint. Finally, the slightly contaminated soil F3 was more similar to the uncontaminated ones, and the uncontaminated soils did not form a single cluster. At the El Gordo Site, the TNT-contaminated soils formed a distinct cluster as well (with 50% similarity, or 72% without sample IN), although soils had been exposed to lower amounts of TNT and for a shorter period of time (Fig. 2b). Finally, the same qualitative shift in community structure was observed in the soil microcosms spiked with increasing amounts of TNT (Fig. 4a, b). In the soil spiked with 140 mg TNT/kg, one bright band appeared late (after 121 days of incubation), whereas in the soils spiked with 1.4 g TNT/kg and 28.5 g TNT/kg, bright bands were already visible after 7 days of incubation. In the soil microcosm spiked with 1.4 g TNT/kg, additional brighter bands appeared after 61 days, whereas the DGGE fingerprint of the most heavily contaminated soil microcosm did not change much after 7 days of incubation. Those changes were reflected in the dendrogram (Fig. 4b). One cluster (49% similarity) contained the samples taken on day 0 in the four microcosms, all samples from the uncontaminated microcosm, and samples from the microcosm spiked with 140 mg TNT/kg until day 14. The other cluster (51% similarity) included one sample (day 61) from the microcosm spiked with 140 mg TNT/kg, and all (but day 0) samples from the microcosm spiked with 1.4 and 28.5 g TNT/kg. One DGGE profile (day 121 in the microcosm spiked with 140 mg TNT/kg) had a unique position in the dendrogram, because of the presence of a bright band that was not or hardly visible in the other DGGE patterns. This band was identified as a very common genus (Pseudomonas) in the heavily TNT-contaminated soil microcosms in the same experiment (see below), indicating that the same bacterial group had been selected under different TNT concentrations. Interestingly, the DGGE patterns of the soil samples collected after 61 and 121 days of incubation in the microcosm spiked with 1.4 g TNT/kg shared a high similarity (81%) with those collected at various time points (but day 0) in the microcosm spiked with 28.5 g TNT/kg, which reveals a convergent adaptation of the microbial community under the selective pressure of high TNT concentration.
Such differences in community composition between uncontaminated and TNT-contaminated soils were reflected in the DGGE patterns of their cultivable community as well (Fig. 3b,c). The cultivable community of the uncontaminated soil REF, although less diverse than its total community (i.e. some dominant cultivable species were detected after 14 days of incubation), was more diverse that the cultivable community of soil KX1. Indeed, the latter displayed a DGGE profile, consisting in a few very bright bands, which was very similar to the pattern of the total community.
All in all, these experiments showed a shift in the total and culturable community structure after TNT contamination. Such results differ from the observations by Ellis et al. [7] in one of the few published studies dealing with the comparative effect of a pollutant on the diversity of the total and the culturable microflora in soils. In their study, metal contamination affected the physiological status of bacteria, so that the number of bacteria capable of responding to laboratory culture and their taxonomic distribution were altered, but did not affect the diversity of the total soil bacterial community.
Dominant species in the total and culturable microflora of TNT-contaminated soils
Bacterial species present in uncontaminated soils from Bourges and El Gordo were not identified, as the DGGE patterns were too diverse to easily excise bands, except for the two bright bands in soil F1 (Fig. 1a) that corresponded to a Pseudomonas sp. and an uncultured bacterium clone (Figs. 5, 6). In the pristine soil used for the spiking experiment with various amounts of TNT, we identified a bacterium probably belonging to the genus Methylobacterium, a Lysobacter sp. and a Ralstonia sp. (Figs. 4a, 6). Regarding the culturable community of uncontaminated soils, a number of numerically dominant species were identified in the uncontaminated soil REF from Bourges: a Pseudomonas sp., an Acinetobacter sp., Variovorax spp., a Rhizobium sp., a Paracoccus sp., and an Achromobacter/Alcaligenes sp. (Figs. 3b, 5, 6). They all belonged to bacterial groups with known culturable representatives. Therefore, we did not identify as-yet uncultured soil bacteria on solid dilute media, like Janssen et al. [25] did by identifying single colonies. These results do not mean that fastidious microorganisms did not grow on the dilute medium, but rather that they did not show up among the dominant species of the DGGE fingerprint. Interestingly, among the CFU from soil REF, various members of the mostly unculturable phylum Acidobacteria could be detected by using Acidobacteria-specific PCR primers, whereas none was detected among the CFU from soil KX1 (George et al., unpublished data).
In the TNT-contaminated soils from Bourges, most bright bands were identified as Pseudomonas spp. (Figs. 1a, 5), and one band corresponding to a Brevundimonas sp. (Figs. 1a, 6) was present in most contaminated soils as well. The same genera made up the cultivable community of the TNT-contaminated soil KX1 (Figs. 3c, 5, 6). Finally, soils KL1 and KL2 harboured additional dominant bands identified as an Acidovorax sp., a Parvibaculum sp. and an uncultured γ-Proteobacterium affiliated to the genus Luteimonas (Figs. 1a, 6). One additional dominant band was identified in soils KF6b and KF6c as a Thermomonas sp. (Figs. 1a, 6). In the TNT-contaminated soils from El Gordo (UEE, IN, C15, J15, C29, and J29), the numerically dominant species were identified as Pseudomonas spp. and Rhodanobacter spp. (Figs. 2a, 5, 6). The artificial contamination of a pristine soil with TNT led to the emergence of species closely related to those found in the contaminated soils from Bourges and El Gordo. In the DGGE pattern of the soil containing 140 mg TNT/kg, a bright band appearing after 121 days of incubation was a Pseudomonas sp. (Figs. 4a, 5). In the DGGE fingerprint of the soil containing 1.4 g TNT/kg, the bright bands appearing at days 7 and 14 corresponded to a Variovorax sp., a Lysobacter sp. and a Pseudomonas sp. (Figs. 4a, 5, 6). After 61 days of incubation, the Pseudomonas sp. band was even brighter, and two additional bands (bands nos. 1 and 10, Fig. 4a) corresponding to another Pseudomonas sp. and a Brevundimonas sp. were detected. Finally, in the heavily contaminated soil (28.5 g TNT/kg), all bright bands were Pseudomonas spp., except for one band (band nos. 12 or 13, Fig. 4a) identified as a Delftia sp. (Fig. 6).
To sum up, most dominant species in the total and culturable bacterial community of the historically TNT-contaminated soils from Bourges and El Gordo and in the artificially TNT-contaminated soil microcosms corresponded to Pseudomonas spp. (Fig. 5), an occurrence previously reported by Fuller and Manning [15] for TNT-contaminated soils using classical cultivation techniques and very recently by Travis et al. [44]. This systematic predominance of pseudomonads in soils impacted with TNT is very likely related to their degradative properties. Several pseudomonads have been isolated from TNT-contaminated environments and were shown to transform TNT to its amino derivatives [9]. However, these amino derivatives tend to accumulate in the environment and are not degraded further. More interesting, a few bacteria have been reported to denitrate TNT [10, 40] with the production of metabolites known to be easily mineralized [34]. Among those, a new Pseudomonas aeruginosa strain was recently isolated by Eyers et al. from the soil KX1 (manuscript submitted).
Interestingly, we also found other dominant bacterial families in most TNT-contaminated soils from Bourges and El Gordo, as well as in the microscosms spiked with TNT: Comamonadaceae (Acidovorax, Variovorax, Delftia genera), Xanthomonadaceae (Thermomonas, Rhodanobacter, Lysobacter genera) and Caulobacteraceae (Brevundimonas genus), and in one case Rhizobiaceae (Parvibaculum genus). They were not present as dominant species in the total and cultivable community of uncontaminated soils. All of them are Gram-negative, which confirms previous observations by Fuller and Manning [15] that Gram-negative bacteria are predominant over Gram-positive bacteria in TNT-contaminated soils. However, the study of these authors was based on phospholipid fatty acid analyses and the identity of the species was not determined. The molecular mechanisms underlying the differential sensitivity of Gram-positive and Gram-negative bacteria to TNT remain to be deciphered.
Most of these bacterial genera have been cited in the literature in relation to the biodegradation of different environmental pollutants, like polycyclic aromatic hydrocarbons (PAHs) [8, 22, 23, 45], trichloroethylene [16], aniline [2], dichloromethane [25], alkylbenzenesulfonates [37], and various pesticides [3, 4, 21, 31, 33, 39]. However, apart from Stenotrophomonas sp. OK-5 (Xanthomonadaceae), which is able to resist TNT-mediated stress by producing stress shock proteins [19] and to grow on TNT as sole nitrogen source [35], no other member of the families Comamonadaceae, Xanthomonadaceae and Caulobacteraceae has been reported so far to metabolize TNT. Indeed, bacteria known to degrade TNT aerobically and/or anaerobically belong to Pseudomonadaceae, Enterobacteriaceae, Mycobacteriaceae, Nocardiaceae, Desulfovibrionaceae, Clostridiaceae and Bacillaceae [36, 40, 41]. The presence of species belonging to Comamonadaceae, Xanthomonadaceae and Caulobacteraceae in soils with very high concentrations of TNT, where pore water is very probably saturated with TNT [44], suggests that those new species are at least tolerant to the presence of TNT. It could also be that among those, some are able to degrade TNT, but this remains to be proved. Nevertheless, the present study opens a field of investigation towards putative new TNT-degrading bacteria.
In conclusion, this work showed that TNT had a significant impact on soil bacterial community structure. To our knowledge, this is the first study providing an extensive characterization of the prokaryotic microflora of soils sampled at different locations and with long-term and short-term TNT contamination, and of a pristine soil artificially contaminated with TNT. Given its adverse impact on the indigenous microbial communities, there is an important need for decontaminating sites chronically polluted with TNT.
References
Anonymous (1972) Soil survey laboratory methods and procedures for collecting soil samples. Soil survey investigations report n°1, U.S. Department of Agriculture, Soil Conservation Service, Washington, DC
Boon N, Goris J, De Vos P, Verstraete W, Top EM (2001) Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl Environ Microbiol 67:1107–1115
Dejonghe W, Berteloot E, Goris J, Boon N, Crul K, Maertens S, Höfte M, De Vos P, Verstraete W, Top EM (2003) Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain. Appl Environ Microbiol 69:1532–1541
Deshpande NM, Sarnaik SS, Paranjpe SA, Kanekar PP (2004) Optimization of dimethoate degradation by Brevundimonas sp. MCM B-427 using factorial design: studies on interactive effects of environmental factors. World J Microbiol Biotechnol 20:455–462
El Fantroussi S, Verschuere L, Verstraete W, Top EM (1999) Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol 65:982–988
El Fantroussi S, Belkacemi M, Top EM, Mahillon J, Naveau H, Agathos SN (1999) Bioaugmentation of a soil bioreactor designed for pilot scale anaerobic bioremediation studies. Environ Sci Technol 33:2992–3001
Ellis RJ, Morgan P, Weightman AJ, Fry JC (2003) Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol 69:3223–3230
Eriksson M, Sodersten E, Yu Z, Dalhammar G, Mohn WW (2003) Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from Northern soils. Appl Environ Microbiol 69:275–284
Esteve-Nunez A, Caballero A, Ramos JL (2001) Biological degradation of 2,4,6-trinitrotoluene. Microbiol Mol Biol Rev 65:335–352
Esteve-Nunez A, Ramos JL (1998) Metabolism of 2,4,6-trinitrotoluene by Pseudomonas sp. JLR11. Environ Sci Technol 32:3802–3808
Eyers L, Stenuit B, El Fantroussi S, Agathos SN (2004) Microbial characterization of TNT-contaminated soils and anaerobic TNT degradation: high and unusual denitration activity. In: Verstraete W (ed) Proceedings of the 5th European symposium on environmental biotechnology. Taylor and Francis Group, London, UK, pp 51–54
Eyers L, Smoot JC, Smoot LM, Bugli C, Urakawa H, McMurry Z, Siripong S, El Fantroussi S, Lambert P, Agathos SN, Stahl DA (2006) Discrimination of shifts in a soil microbial community associated with TNT-contamination using a functional ANOVA of 16S rRNA hybridized to oligonucleotide microarrays. Environ Sci Technol 40:5867–5873
Eyers L, Agathos SN, El Fantroussi S (2004) Denaturing Gradient Gel Electrophoresis as a fingerprinting tool for analyzing communities in contaminated environments. In: Spencer JFT, Ragout de Spencer AL (eds) Environmental microbiology: methods and protocols. Humana Press, Totowa, NJ, pp 407–417
Ferrari BC, Binnerup SJ, Gillings M (2005) Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol 71:8714–8720
Fuller ME, Manning JF (1998) Evidence for differential effects of 2,4,6-trinitrotoluene and other munitions compounds on specific subpopulations of soil microbial communities. Environ Toxicol Chem 17:2185–2195
Futamata H, Nagano Y, Watanabe K, Hiraishi A (2005) Unique kinetic properties of phenol-degrading Variovorax strains responsible for efficient trichloroethylene degradation in a chemostat enrichment culture. Appl Environ Microbiol 71:904–911
Gong P, Gasparrini P, Rho D, Hawari J, Thiboutot S, Ampleman G, Sunahara GI (2000) An in situ respirometric technique to measure pollution-induced microbial community tolerance in soils contaminated with 2,4,6-trinitrotoluene. Ecotox Environ Saf 47:96–103
Hane BG, Jager K, Drexler HG (1993) The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 14:967–972
Ho E-M, Chang H-W, Kim S-I, Kahng H-Y, Oh K-H (2004) Analysis of TNT (2,4,6-trinitrotoluene)-inducible cellular responses and stress shock proteome in Stenotrophomonas sp. OK-5. Curr Microbiol 49:346–352
Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396
Kamagata Y, Fulthorpe RR, Tamura K, Takami H, Forney LJ, Tiedje JM (1997) Pristine environments harbor a new group of oligotrophic 2,4- dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol 63:2266–2272
Kanaly RA, Harayama S, Watanabe K (2002) Rhodanobacter sp strain BPC1 in a benzo[a]pyrene-mineralizing bacterial consortium. Appl Environ Microbiol 68:5826–5833
Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 70:1777–1786
Kilmer VJ, Alexander LT (1949) Methods of making mechanical analysis of soils. Soil Sci 68:15–24
Krausova VI, Robb FT, González JM (2006) Biodegradation of dichloromethane in an estuarine environment. Hydrobiologia 559:77–83
Leadbetter JR (2003) Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Curr Opin Microbiol 6:274–281
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
MacNaughton SJ, Stephen JR, Venosa AD, Davis GA, Chang Y-J, White DC (1999) Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol 65:3566–3574
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Meyers S, Deng S, Basta NT, Clarkson WW, Wilber GG (2007) Long-term explosive contamination in soil: effects on soil microbial community and bioremediation. Soil Sediment Contam 16:61–77
Müller RH, Jorks S, Kleinsteuber S, Babel W (1999) Comamonas acidovorans strain MC1: a new isolate capable of degrading the chiral herbicides dichlorprop and mecoprop and the herbicides 2,4-D and MCPA. Microbiol Res 154:241–246
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nalin R, Simonet P, Vogel TM, Normand P (1999) Rhodanobacter lindaniclasticus gen. nov., sp. nov., a lindane-degrading bacterium. Int J Syst Bacteriol 49:19–23
Nishino SF, Paoli GC, Spain JC (2000) Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl Environ Microbiol 66:2139–2147
Oh KH, Kim YJ (1998) Degradation of explosive 2,4,6-trinitrotoluene by s-triazine degrading bacterium isolated from contaminated soil. Bull Environ Contam Toxicol 61:702–708
Peres CM, Agathos SN (2000) Biodegradation of nitroaromatic pollutants: from pathways to remediation. Biotechnol Ann Rev 6:197–220
Schleheck D, Tindall BJ, Rosselló-Mora R, Cook AM (2004) Parvibaculum lavamentivorans gen. nov., sp. nov., a novel heterotroph that initiates catabolism of linear alkylbenzenesulfonate. Int J Syst Evol Microbiol 54:1489–1497
Siciliano SD, Roy R, Greer CW (2000) Reduction in denitrification activity in field soils exposed to long term contamination by 2,4,6-trinitrotoluene (TNT). FEMS Microbiol Ecol 32:61–68
Smejkal CW, Seymour FA, Burton SK, Lappin-Scott HM (2003) Characterisation of bacterial cultures enriched on the chlorophenoxyalkanoic acid herbicides 4-(2,4-dichlorophenoxy) butyric acid and 4-(4-chloro-2-methylphenoxy) butyric acid. J Ind Microbiol Biotechnol 30:561–567
Stenuit B, Eyers L, Rozenberg R, Habib-Jiwan J-L, Agathos SN (2006) Aerobic growth of Escherichia coli with 2,4,6-trinitrotoluene (TNT) as sole nitrogen source and evidence of TNT denitration by whole cells and cell-free extracts. Appl Environ Microbiol 72:7945–7948
Stenuit B, Eyers L, El Fantroussi S, Agathos SN (2005) Promising strategies for the mineralisation of 2,4,6-trinitrotoluene. Rev Environ Sci Biotechnol 4:39–60
Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA (2004) New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol 70:4748–4755
Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245
Travis ER, Hannink NK, van der Gast CJ, Thompson IP, Rosser SJ, Bruce NC (2007) Impact of transgenic tobacco on trinitrotoluene (TNT) contaminated soil community. Environ Sci Technol 41:5854–5861
Vacca DJ, Bleam WF, Hickey WJ (2005) Isolation of soil bacteria adapted to degrade humic acid-sorbed phenanthrene. Appl Environ Microbiol 71:3797–3805
Wikström P, Andersson A-C, Nygren Y, Sjöström J, Forsman M (2000) Influence of TNT transformation on microbial community structure in four different lake microcosms. J Appl Microbiol 89:302–308
Wilke B-M, Gattinger A, Frölich E, Zelles L, Gong P (2004) Phospholipid fatty acid composition of a 2,4,6-trinitrotoluene contaminated soil and an uncontaminated soil as affected by a humification remediation process. Soil Biol Biochem 36:725–729
Acknowledgements
This work was supported by a contract from the European Commission (QLK3-CT-2001-00345). Isabelle George benefited from a grant of “Chargée de Recherches” of the Fund for Scientific Research of Belgium (F.R.S-FNRS). Tristan Félix, a student intern, is acknowledged for his excellent technical assistance. We thank S. El Fantroussi for helpful discussions, and P. van Dillewijn and J. L. Ramos (CSIC, Granada, Spain) for providing the soil samples from El Gordo.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
George, I., Eyers, L., Stenuit, B. et al. Effect of 2,4,6-trinitrotoluene on soil bacterial communities. J Ind Microbiol Biotechnol 35, 225–236 (2008). https://doi.org/10.1007/s10295-007-0289-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-007-0289-2