Abstract
A tyrosine ammonia-lyase (TAL) enzyme from the photosynthetic bacterium Rhodobacter sphaeroides (RsTAL) was identified, cloned and functionally expressed in Escherichia coli, where conversion of tyrosine to p-hydroxycinnamic acid (pHCA) was demonstrated. The RsTAL enzyme is implicated in production of pHCA, which serves as the cofactor for synthesis of the photoactive yellow protein (PYP) in photosynthetic bacteria. The wild type RsTAL enzyme, while accepting both tyrosine and phenylalanine as substrate, prefers tyrosine, but a serendipitous RsTAL mutant identified during PCR amplification of the RsTAL gene, demonstrates much higher preference for phenylalanine as substrate and deaminates it to produces cinnamic acid. Sequence analysis showed the presence of three mutations: Met4 → Ile, Ile325 → Val and Val409 → Met in this mutant. Sequence comparison with Rhodobacter capsulatus TAL (RcTAL) shows that Val409 is conserved between RcTAL and RsTAL. Two single mutants of RsTAL, Val409 → Met and Val 409 → Ile, generated by site-directed mutagenesis, demonstrate greater preference for phenylalanine compared to the wild type enzyme. Our studies illustrate that relatively minor changes in the primary structure of an ammonia-lyase enzyme can significantly affect its substrate specificity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ammonia-lyases are a family of enzymes that catalyze the deamination of amino acids. Members of this family include phenylalanine ammonia lyase (PAL) that converts phenylalanine to cinnamate (CA) [1, 13, 14], and histidine ammonia lyase (HAL) that converts histidine to urocanic acid [15]. Some PAL enzymes, in addition to phenylalanine, will also accept tyrosine as substrate and are therefore called phenylalanine/tyrosine ammonia-lyase (PAL/TAL) or tyrosine ammonia lyase (TAL) depending on the relative activity towards these substrates. Deamination of tyrosine by TAL produces p-hyroxycinnamic acid (pHCA, also known as p-coumaric acid). All enzymes in the PAL/TAL/HAL family contain a conserved Ala–Ser–Gly amino acid motif that undergoes autocatalytic cyclization to generate a 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO) group. This MIO group acts as the catalytic electrophile that carries out the E1cb-like elimination of ammonia and a non-acidic β-proton from the substrate [6]. This proposed mechanism is supported by studies of the crystal structure of the Pseudomonas putida HAL [15], the R. glutinis PAL [4, 17] as well as the parsley PAL [13] enzymes.
The PAL enzyme from plant sources performs the first reaction of the phenylpropanoid pathway and converts phenylalanine to CA. Further hydroxylation of CA produces pHCA which plays a pivotal role in production of a diverse array of plant secondary metabolites. The presence of PAL/TAL enzymes has been reported in some microorganisms [2, 12, 19], with possible involvement in the biosynthesis of secondary metabolites. However, similar to their plant counterparts, most reported microbial PAL/TAL enzymes prefer phenylalanine as their substrate. Exceptions to this rule are the recent reports that describe the identification, characterization, cloning and functional expression in E. coli of a TAL enzyme from the photosynthetic bacteria, Rhodobacter capsulatus [10], and R. sphaeroides [8, 16]. The R. capsulatus enzyme has 32% identity with the plant PAL sequence of Pinus taeda but lower homology to known yeast PAL/TAL enzymes (e.g. 20% identity with Rhodotorula glutinis PAL). Kyndt et al. suggested involvement of this enzyme in the photosensory system of R. capsulatus based on its location upstream of the pyp gene. The photosensory system of some halophilic purple sulfur bacteria such as Ectothiorhospira halophila, Rhodospirillum salexigens and Chromatium salexigens consists of a small 14 kDa water-soluble protein designated photoactive yellow protein (PYP). The presence of a covalently bound pHCA, which is the chromophore of this protein, and its photoisomerization from trans to cis configuration during the photocycle [7, 9, 20] underlines a potential physiological role for this chemical in photosynthetic bacteria.
Our interest in PAL/TAL enzymes stems from their involvement in conversion of aromatic amino acids phenylalanine and tyrosine to CA and pHCA, respectively. These compounds are of interest due to their potential as starting materials for synthesis of a wide array of chemicals including flavors, fragrances, pharmaceuticals, biocosmetics, and health and nutrition products. Herein, we report on identification, cloning and expression in E. coli, followed by its biochemical characterization, of a TAL enzyme from R. sphaeroides (RsTAL). We further describe identification of Val409 as a key residue in RsTAL that determines its substrate specificity. Mutations of Val409 to either methionine or isoleucine generated mutant enzymes with greater preference for phenylalanine as substrate compared to the wild type enzyme.
Materials and methods
Plasmids and strains
Plasmids pKK223-3 was obtained from Pharmacia (GE Healthcare, Piscataway, NJ). R. sphaeroides genomic DNA was obtained from American Type Culture collection (ATCC, Menasses, VA).
Cloning and expression of RcTAL from R. capsulatus and RsTAL from R. sphaeroides
The RcTAL gene was cloned and expressed under the control of a tac promoter in vector pKK223-3 in the E. coli BL21(DE3) strain (Invitrogen, Carlsband, CA) as described by Kyndt et al. [10]. The RsTAL gene (Genbank accession # ZP 00005404) was PCR amplified using TaKaRa DNA polymerase (TaKaRa Mirus Bio, Madison, MI) from R. sphaeroides genomic DNA with the 5′ primer of 5′-CCAACCGTGAAGACGGAATTCATGAAGCCAATGCTCGCCAT-3′ (containing an BbsI site which gives an EcoRI compatible overhang upon digestion, the start codon is underlined) and 3′ primer of 5′-GGACCCTGAAGC TT AGCTGATCGCCATCGAGGTC-3′ (containing a HindIII site, with the stop codon underlined). Plasmid for RsTAL expression was constructed by ligating the EcoRI and HindIII digested PCR fragment into vector pKK223-3 to give plasmid pKK223.RsTAL2, thus allowing RsTAL gene to be expressed under the tac promoter. The plasmid was verified by sequence analysis and expressed in strain BL21(DE3)RP codon plus (Stratagene, San Diego, CA). Sequence alignment of TAL and other ammonia lyases is generated with Vector NTI (Invitrogen).
RsTAL and RcTAL enzyme purification
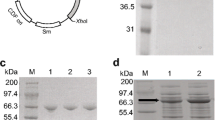
A 500 ml culture of E. coli BL21(DE3)RP (pKK223.RsTAL2) containing RsTAL gene was grown in the Luria Broth (LB) medium plus ampicillin (100 μg/ml), induced with isopropyl-β-d-thiogalactopyranoside (IPTG, 1.0 mM) at the mid log growth phase (OD600=0.5) and incubated (250 rpm) overnight at 37°C. The cells were harvested (2,000×g, 20 min, 4°C) and the pellet resuspended in 10 ml buffer A (Tris–HCl, 50 mM, pH 8.5, DTT, 5.0 mM and tyrosine, 1.0 mM). Protease inhibitor (1.5 EDTA-free tablets per 10 ml) (Roche Biosciences, Palo Alto, CA), was added and cells were passed through the French Pressure Cell twice at 18,000–20,000 psi and centrifuged (18,000×g, 20 min, 4°C) and the supernatant was precipitated at 30% (NH4)2SO4 saturation. Following centrifugation, the supernatant was brought to 50% (NH4)2SO4 saturation, the pellet re-dissolved in buffer A (3.0 ml), and passed through a BioRad 10DG desalting column (BioRad Laboratories, Hercules, CA) before being applied to a 0.8 ml HQ anion exchange perfusion chromatography column (Quaternized polyethyleneimine, POROS® HQ 10 μm Column, PEEK™, 4.6 mm × 50 mm, Applied Biosystems, Foster City, CA). Tyrosine (1.0 mM) was added to both the cell lysate and purification buffer to allow adherence of the enzyme to the column. The column was eluted with a 0–1.0 M NaCl gradient in a buffer system containing Tris-HCl (50 mM, pH 8.5), DTT (5.0 mM) and tyrosine (1.0 mM). The TAL activity in eluted fractions was determined spectrophotometrically and later analyzed by SDS-PAGE electrophoresis. The same procedure was used for purification of E. coli cells containing R. capsulatus TAL enzyme.
Site-directed mutagenesis of RsTAL
The RsTAL mutants, V409M and V409I, were generated by site-directed mutagenesis using the QuickChange Site-directed mutagenesis kit as directed by the manufacturer (Stratagene). The mutations were confirmed by sequence analysis on an Applied Biosystems 3130 DNA sequencing machine.
Protein gel electrophoresis
The purity of the protein samples (4.0 μg of protein per lane) were examined by SDS-PAGE analysis using the 4–12% gradient gel (Invitrogen) and stained with Coomassie blue. Molecular weight marker (Mark 12) was used as the standard (Invitrogen).
PAL/TAL enzyme assays
Spectrophotometric assay
The TAL activity in cell extracts was determined spectrophotometrically as described by Abell and Shen [1]. Initial rates were measured using tyrosine (from 0.01 to 10 mM) in Tris–HCl (50 mM, pH 8.5) buffer and pHCA formation was monitored at 315 nm for 5 min at 25°C. The enzyme activity was calculated using a molar extinction coefficient of 10,000 M−1 cm−1 for pHCA. The PAL activity was similarly measured, by monitoring CA formation at 290 nm using a molar extinction coefficient of 9,000 M−1 cm−1. The protein concentration was measured according to Bradford [3] using bovine serum albumin as the standard.
HPLC assay
Typically the reaction was initiated by adding tyrosine (1.0 mM) to a 1.0 ml solution containing 25 μg purified RsTAL enzyme in Tris-HCl (50 mM, pH 8.5) buffer. An aliquot (50 μl) was removed at various time intervals, and heated to 85°C for 10 min to stop the reaction. The pHCA concentration in each sample was determined by HPLC.
HPLC analysis
E. coli strains transformed with TAL expression vectors were grown and induced in the LB medium. The clarified culture supernatants (800 μl) were filtered (0.2 micron nylon Spin-X spin filter) (Corning, Acton, MS) in a microcentrifuge (5.0 min, 15,000×g, at room temperature) and analyzed for pHCA and CA concentrations by HPLC analysis using a Zorbax SB-C18 column in an Agilent 1100 chromatography system (Agilent technologies, Palo Alto, CA). The solvent system used consisted of a gradient from 5 to 80% acetonitrile containing 0.5% trifluoroacetic acid (TFA) for 8 min, followed by 80% acetonitrile containing 0.5% TFA for 2 min. pHCA and CA were detected at 312 and 278 nm with retention times of 5.4 and 7.1 min, respectively.
Growth and product analysis of RsTAL mutant clones
E. coli cells containing either the wild type or the mutants of RsTAL were grown (37°C, 250 rpm) in 10 ml of LB medium containing 100 μg/ml ampicillin to mid log phase, induced with IPTG (1.0 mM) and incubated (250 rpm) overnight at 37°C. The cells were harvested (2,000×g, 5 min) and the supernatant was clarified by passing through a 0.2 micron nylon filter. The concentrations of pHCA and CA in the culture supernatant was determined by HPLC and the TAL/PAL activity ratio calculated.
Results
Cloning and expression of RcTAL and the putative TAL gene from R. sphaeroides
The GC rich RsTAL gene [8] was expressed as a soluble protein from plasmid pK223.RsTAL2 under the tac promoter in E. coli BL21(DE3) RP codon plus strain. The RsTAL enzyme was purified to near homogeneity following ammonium sulfate fractionation and anion exchange chromatography. In order to compare the enzymatic properties of this enzyme with the previously characterized RcTAL enzyme [10], we cloned and expressed RcTAL under the tac promoter in E. coli and purified the enzyme as described by Kyndt et al. [10].
Comparison of kinetic properties of RsTAL and RcTAL
We studied the kinetic parameters of both purified RsTAL and RcTAL recombinant enzymes and compared them with other known PAL/TAL enzymes (Table 1). RsTAL showed K cat and K m values of 0.02 s−1 and 60 μM while these values for RcTAL were 0.06 s−1 and 160 μM, respectively. Varying the reaction temperatures between 25 and 55°C did not result in improvement of either enzyme’s activities. In addition, the RsTAL activity was measured in an end-point assay using HPLC (see “Materials and methods”) to determine the concentration of pHCA formed (Fig. 1). The rate of pHCA formation during the first minute was significantly higher (5.0 μM pHCA produced) compared to the much slower rate (∼1.0 μM/min) thereafter. Therefore, HPLC analysis suggested that the initial rate of the reaction (0.0–1.0 min) was at least 0.2 s−1, and the steady state rate after 1.0 min was only 0.03 s−1. Kinetic analysis of RsTAL, in the presence of varying concentrations of pHCA, showed an inhibition constant (K i ) of approximately 1.3 μM.
Identification and analysis of RsTAL mutants
During PCR amplification of the RsTAL gene from the genomic DNA, we identified a mutant clone with improved PAL activity compared to the wild type enzyme. The E. coli host TOP10 (Invitrogen) harboring this mutant clone was named strain DPD5077. While the ratio of pHCA/CA formed by the wild type RsTAL enzyme was 29.0, this ratio was 1.7 for the mutant (Table 2). It should be noted that the LB medium used for growth of the E. coli strain expressing the PAL/TAL enzyme contains both phenylalanine and tyrosine, which could be converted to CA and pHCA by this enzyme hence verifying the enzyme’s functional expression.
Sequence analysis showed presence of three mutations: M4I, I325V and V409M in the mutant TAL. Comparison of RsTAL and RcTAL sequences showed that only Val409 was conserved between the two proteins. Sequence alignment of TALs with other PAL and HAL enzymes (Fig. 2) shows that the corresponding residue of Val409, e.g., Ile472 (of R. glutinis PAL), is conserved amongst all PAL enzymes.
Sequence alignment of a selected group of PAL, TAL and HAL enzymes. Generated with Vector NTI. Residues identical among different proteins are highlighted in dark grey, homologous residues and blocks of similar residues are highlighted in light grey. The residue in each protein that aligns with V409 of RsTAL is highlighted in a black frame
We used site-directed mutagenesis to investigate the significance of the point mutation V409M in RsTAL substrate specificity. In addition, we generated V409I mutant, to test whether Ile409 could lead to a similar change of specificity. E. coli cells containing RsTAL mutants were induced and grown as described in “Materials and methods”. Cultures expressing the mutant enzymes produced both CA and pHCA. While the ratio of pHCA to CA produced by wild type RsTAL was 29.0, this ratio was 1.7 for the M4I-I325V-V409M triple mutant and only 0.7 and 0.8 with single mutants V409M and V409I, respectively (Table 2).
Discussion
Unlike some other PAL/TAL enzymes that prefer phenylalanine versus tyrosine as substrate, the Rhodobacter enzymes prefer tyrosine and can therefore be used as tools for understanding the substrate recognition mechanisms of the phenylalanine/tyrosine/histidine ammonia lyase family of enzymes. We identified, cloned and expressed in E. coli the TAL enzyme of R. sphaeroides [8].
The presence of the TAL enzyme in this organism is consistent with the enzyme’s role in pHCA biosynthesis, which acts as a cofactor for PYP function in photosynthetic bacteria [7, 20]. While the catalytic activities of both RcTAL (K cat 0.06 s−1) and RsTAL (K cat 0.02 s−1) enzymes are relatively low, they are probably enough for production of the required levels of pHCA as the cofactor for PYP synthesis. In fact, a highly active TAL enzyme would be detrimental to the host cell, because it would divert the intracellular pool of tyrosine thereby reducing its availability for protein synthesis. In addition, high concentrations (e.g., 10 g/l) of pHCA cause cell death to E. coli (Authors’ unpublished observations). A TAL enzyme with relatively low activity therefore would be more beneficial to the cell’s function.
Current reports on the TAL activities of RcTAL [10] and RsTAL [11, 18] describe activities significantly higher than those observed by us during our studies. The differences in observed rates probably reflect differences in assay conditions used for these determinations. For example, as indicated in the “Results”, during TAL activity measurements using an HPLC method (Fig. 1), the initial rate of the reaction was 0.2 s−1 from 0.0 to 1.0 min followed by the steady state rate of 0.03 s−1 after 1.0 min. This finding is not surprising since pHCA is a strong competitive inhibitor of the enzyme. In addition, this lower measured rate is probably due to the rate of the release of products (pHCA and ammonia) from the TAL enzyme’s active site. In a typical TAL assay measured spectrophotometrically, the assay is usually performed for 3–5 min and the initial high rate of activity, if any, is ignored. The reported rate, therefore, is much lower than the initial observed rate. One could therefore speculate that in assays performed by Kyndt et al. and Watts et al. product release was more rapid under the conditions of their assays. One possible reason for this is the presence of His-tag enzyme used by Watts et al. [18] that could have led to a faster product release from the enzyme. Further detailed studies using stop-flow measurements are required to determine these enzymes’ kinetics unequivocally.
We identified a spontaneous RsTAL mutant during the PCR amplification of the RsTAL gene which was probably introduced by point mutations generated by the non proof-reading DNA polymerase used in the PCR reaction. While the wild type enzyme prefers tyrosine as substrate and predominately produces pHCA, this mutant enzyme (expressed in strain DPD5077) has higher affinity for phenylalanine as substrate and therefore produces similar amounts of pHCA and CA. Upon sequence comparison with other ammonia-lyases, we found that V409M is conserved between RcTAL and RsTAL, and that Val409 aligns with Ile472 in R. glutinis PAL which is a conserved residue among all PALs.
Recently, the crystal structure of RsTAL was determined [11]. In this structure, Val409 is located in the active site in close proximity to the aromatic ring of the substrate. Interestingly, Val409 is positioned at the opposite side of the aromatic ring compared to His89 which is proposed by Louie et al. [11] and Watts et al. [18] to play an important role in differentiating between tyrosine and phenylalanine as substrates. In their studies [11, 18], H89F mutation led to a switch of substrate specificity from tyrosine to phenylalanine. In our current study, mutation of Val409 to either methionine or isoleucine, both of which have longer hydrophobic side chains than valine, allowed the enzyme to recognize both phenylalanine and tyrosine as substrates. Thus it appears likely that in addition to His89, Val409 in the wild type RsTAL enzyme also plays an important role in the preferential binding of tyrosine versus phenylalanine. We speculate that either the H89F mutant or the V409I and V409M mutants may have generated a more favorable hydrophobic interaction with the aromatic side chain of phenylalanine.
In this study, we have provided another example in which relatively minor changes in the primary structure of an ammonia-lyase enzyme could significantly affect its substrate specificity. It is likely that the kinetic properties of the TAL enzymes could be improved through protein engineering approaches such as gene shuffling with the closely related HAL enzymes, which have very high activities. Such engineered TAL enzymes could provide useful catalysts for bioconversion of tyrosine to the industrially attractive pHCA molecule.
References
Abell CW, Shen RS (1987) Phenylalanine ammonia-lyase from yeast Rhodotorula glutinis. Methods Enzymol 142:242–248
Berner M, Krug D, Bihlmaier C, Vente A, Muller R, Bechthold A (2006) Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol 188:2666–2673
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Calabrese J, Jordan DB, Boodhoo A, Sariaslani S, Vannelli T (2004) Crystal structure of phenylalanine ammonia lyase from Rhototorula glutinis. Biochemistry 43:11403–11416
Gatenby AA, Sariaslani FS, Tang X, Qi WW, Vanelli T (2002) Bioproduction of hydroxycinnamic acid. US Patent 6368837
Hermes JD, Weiss PM Cleland WW (1985) Use of nitrogen-15 and detuerium isotope effects to determine the chemical mechanism of phenylalanine ammonia-lyase. Biochemistry 24:2959–2967
Hoff WD, Dux P, Hard K, Devreese B, Nugteren-Roodzat I M, Crielaard W, Boelens R, Kaptein R, Van Beeumen J, Hellingwerf KJ (1994) Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry 33:13959–13962
Huang L, Xue Z (2006) DNA and amino acid sequence of a tyrosine ammonia lyase from the bacterium Rhodobacter sphaeroides. US Patent No. 7,067,302
Kort R, Vonk H, Xu X, Hoff WD, Crielaard W, Hellingwerf KJ (1996) Evidence for trans-cis isomerization of the p-coumaric acid chromophore as the photochemical basis of the photocycle of photoactive yellow protein. FEBS Lett 382:73–78
Kyndt JA, Meyer TE, Cusanovich MA, Meyer JJ, Van Beeumen JJ (2002) Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett 512:240–244
Louie GV, Bowman ME, Moffitt MC, Baiga TJ, Moore BS, Noel JP (2006) Structural determinants and modulation of substrate specificity in phenylalanine–tyrosine ammonia lyases. Chem Biol 13:1327–1338
Moffitt MC, Louie GV, Bowman ME, Pence J, Noel JP, Moore BS (2007) Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry (published online). doi:10.1021/bi061774g
Ritter H, Schulz GE (2004) Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 16:3426–3436
Rosler J, Krekel N, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113:175–179
Schwede TF, Retey J, Schulz GE (1999) Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38:5355–5361
Watts KT, Lee PC, Schmidt-Dannert C (2004) Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem 5:500–507
Wang L, Gamez A, Sarkissian CN, Straub M, Patch MG, Han GW, Striepeke S, Fitzpatrick P, Scriver CR, Stevens RC (2005) Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria. Mol Gen Metab 86:134–140
Watts KT, Mijts BN, Lee PC, Manning AJ, schmidt-Dannert C (2006) Discovery of a substrated selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem Biol 13:1317–1326
Xiang L, Moore BS (2002) Inactivation, complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J Biol Chem 277:32505–32509
Xie A, Hoff WD, Kroon AR, Hellingwerf KJ (1996) Glu46 donates a proton to the 4-hydroxycinnamate anion chromophore during the photocycle of photoactive yellow protein. Biochemistry 35:14671–14678
Acknowledgment
We thank Joseph Calabrese, and Anthony Gatenby for helpful discussions and review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xue, Z., McCluskey, M., Cantera, K. et al. Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from the photosynthetic bacterium Rhodobacter sphaeroides . J Ind Microbiol Biotechnol 34, 599–604 (2007). https://doi.org/10.1007/s10295-007-0229-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-007-0229-1