Abstract
Although machine learning (ML) has made significant improvements in radiology, few algorithms have been integrated into clinical radiology workflow. Complex radiology IT environments and Picture Archiving and Communication System (PACS) pose unique challenges in creating a practical ML schema. However, clinical integration and testing are critical to ensuring the safety and accuracy of ML algorithms. This study aims to propose, develop, and demonstrate a simple, efficient, and understandable hardware and software system for integrating ML models into the standard radiology workflow and PACS that can serve as a framework for testing ML algorithms. A Digital Imaging and Communications in Medicine/Graphics Processing Unit (DICOM/GPU) server and software pipeline was established at a metropolitan county hospital intranet to demonstrate clinical integration of ML algorithms in radiology. A clinical ML integration schema, agnostic to the hospital IT system and specific ML models/frameworks, was implemented and tested with a breast density classification algorithm and prospectively evaluated for time delays using 100 digital 2D mammograms. An open-source clinical ML integration schema was successfully implemented and demonstrated. This schema allows for simple uploading of custom ML models. With the proposed setup, the ML pipeline took an average of 26.52 s per second to process a batch of 100 studies. The most significant processing time delays were noted in model load and study stability times. The code is made available at “http://bit.ly/2Z121hX”. We demonstrated the feasibility to deploy and utilize ML models in radiology without disrupting existing radiology workflow.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Machine learning (ML) has made significant advances in radiology, especially with the applications of artificial neural networks to various medical imaging modalities. However, very few of these algorithms have been integrated into the clinical radiology workflow [1]. ML algorithms have demonstrated promising performance on a variety of tasks, such as Alzheimer’s prediction, mammographic risk scoring, tomographic segmentation, and arthritic joint and muscle tissue segmentation [2,3,4,5,6]. Given a two-fold increase in workload seen by radiologists from 1999 to 2010 [7], ML algorithms have the potential to help radiologists manage this burden and improve performance.
Unfortunately, the impact of these algorithms has largely been limited to pilot experiments and technical discussions in literature to date. Complex radiology IT system and associated Picture Archiving and Communication System (PACS) in actual clinical radiology practice, along with the relative inaccessibility of these systems to outside industry and academic ML engineers, pose unique challenges in creating an efficient, simple, and practical ML algorithm clinical integration schema [8]. Accordingly, the 2018 RSNA Artificial Intelligence Summit emphasized that developing systems to deploy ML algorithms in clinical practice is now an essential component in improving algorithm quality and radiology performance [9].
Several industry teams have developed custom ML-focused PACS, but integrating these systems will require extensive trial and error. Moreover, implementing new PACS is in itself a challenging task, often requiring retraining staff, disrupting radiology workflow, and potentially raising data security concerns with proprietary code integrating with hospital IT [10]. Therefore, we proposed, developed, and demonstrated a simple and efficient system for integrating ML models into the standard radiology workflow and existing PACS that can serve as the basis for testing homegrown or commercial algorithms.
Methods
DICOM/GPU Server
This institutional review board approved, written informed consent waived, and HIPAA-compliant study involved the establishment of a DICOM/GPU server and software pipeline at a metropolitan, academic county hospital intranet to demonstrate proof-of-concept of clinical integration of ML algorithms in radiology. A free, open-source, and lightweight Orthanc DICOM server (Orthanc 1.0; University Hospital of Liége, Belgium) was installed onto the DICOM/GPU server with a Linux operating system (Ubuntu 16.04; Canonical, London, England). This server has a six-core AMD Ryzen 5 2600 processor (clock speed at 3.4 GHz) (AMD, Santa Clara, Calif), 16 GB of DDR4 SDRAM, and an NVIDIA Titan V Volta graphical processing unit (Nvidia Corporation, Santa Clara, Calif) with CUDA 9.2 and CuDNN 7.5 (Nvidia).
The DICOM/GPU server was placed within the intranet of the hospital’s radiology IT network and set up to communicate with the hospital PACS. The DICOM/GPU server was designated an IP address on the PACS virtual routing and forwarding protocol, which was assigned to the MAC address of the server, to minimize security and firewall policies if the server resides on another subnetwork. A static port configuration on a fabric extender (FEX) switch, which was also connected to the PACS servers, was established in the hospital datacenter. The cluster of servers were connected with a pair of redundant 10 GB copper connections that correspond to a pair of redundant FEX switches for business continuity. The DICOM/GPU server was set up with a single 1 GB connection, which can be swapped with redundant 10 GB or 4 × 1 GB network cards to handle increased volume to the DICOM/GPU server. The Orthanc server was configured by specifying the PACS alias, IP address, ports and Application Entity Titles, and a vendor patch parameter in the configuration file [8].
Clinical ML Integration
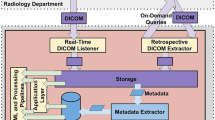
In the standard data flow, patient imaging data moves from the medical imaging equipment to the hospital PACS (Fig. 1.1), where images can then be queried from the radiology workstations (Fig. 1.2). The DICOM/GPU server expands the data flow to incorporate ML algorithms within a standard PACS environment. Radiologists can transmit studies from PACS to the Orthanc DICOM server (Fig. 1.3). The GPU server watches for new instances in Orthanc and sends DICOMs to appropriate ML models, based on DICOM headers (Fig. 1.4). Additionally, radiologists can connect to a virtual session in the GPU server to choose to apply a specific algorithm. The GPU server transforms outputs into a new series (Fig. 1.5) and uploads back to the Orthanc DICOM server (Fig. 1.6). The GPU server transmits new ML series back to the hospital PACS (Fig. 1.7). All actions by the GPU server to process and modify DICOM images are conducted via Python code. A live demo was conducted with a BI-RADS breast density classification algorithm. One hundred digital screening mammograms from April 1 to April 12, 2019, were transmitted from a PACS workstation to the DICOM/GPU server to test implementation and analyze time delays greater than 0.5 s in the ML pipeline.
The standard flow of medical imaging data can be expanded to include the input of machine learning models. (1) Medical imaging equipment transfers study to PACS. (2) PACS sends images to radiologist workstations. (3) Studies from Hospital PACS can be manually or automatically transmitted to the Orthanc DICOM server, situated within the combined DICOM/GPU server. (4) DICOM server watches for new instances in Orthanc and sends DICOMs to appropriate ML models, based on DICOM headers. (5) GPU server transforms outputs into a new DICOM series and (6) uploads back to the Orthanc DICOM server. (7) GPU server, either automatically or manually, transmits new ML series back to hospital PACS
ML Model Updates and Intervention
This clinical ML integration was designed to be modular to allow the ML engineer to easily add or remove ML models and audit ML outputs. The workflow has three intervention points: DICOM headers, the deployed ML models, and the Orthanc server itself (Fig. 2). The DICOM headers (Study Description, Body Part Examined, Modality, and Series Description) are used to determine to which ML model to apply to the study (Fig. 2.1). These ML models can be custom, pretrained models or industry acquired models (Fig. 2.2). The Orthanc server can be inspected by the ML supervisor to perform a quality assurance check on the ML models (Fig. 2.3).
Hospital ML administrators and radiologists have 3 key points to assess and intervene in the ML workflow. (1) They can determine which algorithms will be applied to which DICOM series by modifying body part, modality, and series description that should be identified in the DICOM header information; (2) upload new ML pretrained algorithms to be integrated into the clinical workflow; and (3) audit the DICOM server to perform quality assurance checks
Results
A Live Demo of Breast Density Classification
As a proof of concept and live demonstration, the clinical ML integration was tested with a breast density classification model. This model was integrated into the ML workflow by uploading only two files to the DICOM/GPU server: an h5 file containing the model weights and architecture and a Python script implementing preprocessing, prediction, and postprocessing steps. The model was used to test the integration system and any time delays greater than 0.50 s in the ML integration pipeline. The code is made available at http://bit.ly/2Z121hX.
Machine Learning Model Description
Studies were identified as candidates for the breast density classification model using DICOM header metadata: Study Description, Series Description, Modality, and Body Part Examined. Candidate selection and DICOM image preprocessing steps are institution- and algorithm-specific parameters. The DICOM/GPU server did not fail to recognize any studies when DICOM parsing header metadata. Our model used Xception architecture to classify the breast density of 2D mammography images using the defined BI-RADS classifications [11, 12]. This model predicted a probability for each BI-RADS breast density class (A, B, C, or D) for each 2D mammogram image within the exam, which typically consisted of two views of each breast for a total of four images. This model averaged the probabilities to predict an output. The detailed description of the ML model is outside the scope of this manuscript, since the described model is diagnosis and team specific, and can be substituted with a different model.
The model then generated a new DICOM file with an image detailing the model and its predictions (Fig. 3). This DICOM is appended as a new series, titled “ML_models” within the same study. The outputs of future ML models applied to this study would be appended to this series to avoid disrupting existing workflows and allow the radiologist to incorporate model findings at their discretion.
Output of BI-RADS density classification algorithm via clinical ML pipeline integrated with PACS. Study was transmitted from PACS workstation to DICOM/GPU server for analysis. Output was generated as a new series, “ML_models,” and transmitted back to PACS. Output DICOM originally appeared as a thumbnail in left sidebar without interfering with mammography hanging protocols
ML Server Times Delays
Significant time delays in clinical ML integration can reduce its utility in certain time-sensitive cases. Although the exact time delay will depend on compute power, model architecture, and network speed, measured times in our set up were reported to provide an estimate. Table 1 lists delays greater than 0.50 s in processing time on the DICOM/GPU server. The mean time required to analyze one study was 86.27, 32.45, and 26.52 s when processing studies in batch of 1, 10, and 100 studies, respectively. The batch size affected the study stability time, which is the time required to ensure no new DICOMs will be loaded into Orthanc as part of the study. Orthanc defaulted to waiting 60 s for new DICOM files for a specific study, but this can be configured as necessary. Because Orthanc received studies in parallel, batching studies led to smaller mean delay times. Study stability time was recorded as 60.83, 6.34, and 0.62 s for batch sizes of 1, 10, and 100 studies, respectively. The Xception model load time, the time required to load the Xception architecture into GPU memory, averaged 26.52 s. The instance analysis time, the time for the server to read, preprocess, and output a prediction, averaged 0.91 s. Model load time and study stability time were the bottlenecks in this workflow.
Discussion
We developed a simple, efficient, and open-source pipeline for integrating ML algorithms into clinical radiology workflow using a DICOM/GPU server that minimizes disruption to the existing PACS system and radiologists’ diagnostic work. This pipeline, which is agnostic to hospital IT systems, would allow for easy supervision, modification, and analysis by radiologist and radiology IT administrators to both prospectively and retrospectively monitor the performance and utility of ML algorithms.
Current challenges of deploying ML models in existing IT infrastructures have limited the utility of ML in clinical radiology workflow. NVIDIA released the Clara SDK in 2019, which allows the engineers to deploy ML radiology algorithms in the clinic. This SDK offers a parallel implementation of clinical ML integration and is a positive step for clinical ML integration (NVIDIA Clara, 2019). While the library offers advanced features and GUIs, it further abstracts the ML integration implementation, thus adding complexity for complex use cases that may not follow traditional ML pipelines, and relies on NVIDIA for maintenance and upkeep. Existing literature on ML studies that have prospectively analyzed models in radiology often involved third-party software to display the results or adopted outside infrastructure into the clinical data center [13,14,15]. This could affect clinical workflow and create barriers for researchers to properly evaluate the algorithms. Furthermore, implementation of outside industry hardware and software at the core of clinical radiology data center, with little control by the department, could raise concerns over data security and patient confidentiality. By open-sourcing the clinical ML integration architecture, we enabled radiologists and PACS engineers to integrate existing algorithms into the workflow, identify new opportunities to utilize ML in radiology, and test ML models in real world environments while retaining full control of the environment and ML models.
In clinical practice, constant evaluation of model accuracy and shortcomings, in conjunction with model calibration and fine-tuning, is necessary to arrive at the best possible solution. Misclassification and measurement errors will inevitably exist in ML models, and both over-reliance on ML models and complete mistrust/avoidance of them can both reduce the effectiveness of radiologists [16]. We support the implementation of quality assurance (QA) systems when deploying ML models and suggest a three-pronged approach for QA (Fig. 4). Radiologists who notice a mislabeled instance may access a survey or webform to report misclassifications (Fig. 4.1). Additionally, certain PACS viewers can be configured to embed radiology annotations in DICOM headers and transmit them back to the DICOM/GPU server [17]. Radiologists can directly correct the annotation and send the correction back to the Orthanc/GPU server (Fig. 4.2). Embedding annotations, if possible with given PACS, can be an efficient QA mechanism because it reduces the number of steps required for a radiologist to provide feedback. Lastly, existing QA checks can also catch ML output mistakes (Fig. 4.3). This three-pronged QA framework can be integrated with a future schema to retrain the ML models and improve performance.
Three-pronged framework for performing quality assurance (QA) check on an incorrect output by ML model. (1) Radiologist enters patient accession number and model name into intranet webform. (2) Radiologist makes text and/or visual annotations in a PACS viewer and saves this information to DICOM header with corrections. Radiologist transmits back to DICOM/GPU Server, which reads annotations and notes incorrect ML output. (3) The hospital’s standard radiology QA protocol catches the ML output error. All identified outputs are stored in an ML QA database
Loading the ML model at the start of a study rather than before each instance reduced delay times significantly. When scaling a successful, high-impact model, a dedicated DICOM/GPU server with the ML model preloaded can be established to process those DICOM images. Study stability time can be reduced by having the DICOM/GPU server automatically query PACS for new studies at fixed time intervals. Studies would be analyzed prior to radiologists opening the study, and batched queries would increase the batch size and reduce delay times.
Our study has several limitations. First, our approach requires extra storage because it sends an additional DICOM image to the existing PACS infrastructure. However, the extra DICOM image allows vendor-neutral integration and becomes easily visible to the radiologist. Second, our demo reads the DICOM header to determine the most appropriate ML algorithm, and this parameter must be tailored to each individual institution’s protocol.
Conclusion
Overall, our study provides a technical framework for deploying ML algorithms in radiology and tested this framework at our host institution with a breast density classification algorithm. This framework is modular, open-source, and vendor agnostic, allowing it to be adapted for a wide variety of ML applications in radiology. Furthermore, QA checks are essential to ensuring an ML model’s clinical performance, and our framework provides the flexibility for researchers to implement model-specific and hospital-specific QA systems.
References
Choy G, Khalilzadeh O, Michalski M, et al: Current Applications and Future Impact of Machine Learning in Radiology. Radiology 288(2):318–328,2018.
Ding Y, Sohn JH, Kawczynski MG, et al: A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 290(2):456–464,2018.
Kallenberg M, Petersen K, Nielsen M, et al: Unsupervised Deep Learning Applied to Breast Density Segmentation and Mammographic Risk Scoring. IEEE Trans Med Imaging 35(5):1322–1331,2016.
Polan DF, Brady SL, Kaufman RA: Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys Med Biol 61(17):6553–6569,2016.
Pedoia V, Majumdar S, Link TM: Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29(2):207–221,2016.
Bickelhaupt S, Paech D, Kickingereder P, et al: Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging 46(2):604–616,2017.
McDonald RJ, Schwartz KM, Eckel LJ, et al: The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad Radiol 22(9):1191–1198,2015.
Jodogne S: The Orthanc Ecosystem for Medical Imaging. J Digit Imaging 31(3):341–352,2018.
Chokshi FH, Flanders AE, Prevedello LM, Langlotz CP: Fostering a Healthy AI Ecosystem for Radiology: Conclusions of the 2018 RSNA Summit on AI in Radiology. Radiol Artif Intell 1(2):190021,2019.
Berkowitz SJ, Wei JL, Halabi S: Migrating to the Modern PACS: Challenges and Opportunities. RadioGraphics 38(6):1761–1772,2018.
Sickles E, Bassett L, et al: ACR BI-RADS® Mammography. American College of Radiology; 2013. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Bi-Rads/.
Chollet F: Xception: Deep Learning with Depthwise Separable Convolutions. ArXiv 161002357(Cs),2016;http://arxiv.org/abs/1610.02357. Accessed July 1, 2019.
Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC: Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. Npj Digit Med 1(1):39,2018.
Kanagasingam Y, Xiao D, Vignarajan J, Preetham A, Tay-Kearney M-L, Mehrotra A: Evaluation of Artificial Intelligence–Based Grading of Diabetic Retinopathy in Primary Care. JAMA Netw Open 1(5):e182665,2018.
Topol EJ: High-performance medicine: the convergence of human and artificial intelligence. Nat Med 25(1):44,2019.
Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G: Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern Med 178(11):1544–1547,2018.
Heydari A, Lituiev D, Vu TH, Seo Y, Sohn JH: A DICOM-embedded Annotation System for 3D Cross-sectional Imaging Data. Abstr Radiol Soc N Am,2019.
Acknowledgments
We thank the NVIDIA corporation for the donation of NVIDIA-Titan Xp GPU.
Funding
Jae Ho Sohn was supported by the National Institute of Biomedical Imaging and Bioengineering T32-EB001631 grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sohn, J., Chillakuru, Y.R., Lee, S. et al. An Open-Source, Vender Agnostic Hardware and Software Pipeline for Integration of Artificial Intelligence in Radiology Workflow. J Digit Imaging 33, 1041–1046 (2020). https://doi.org/10.1007/s10278-020-00348-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00348-8