Abstract
Recent healthcare policies have influenced the manner in which patient data is handled in research projects, and the regulations concerning protected health information have become significantly tighter. Thus, new procedures are needed to facilitate research while protecting the confidentiality of patient data and ensuring the integrity of clinical work in the expanding environment of electronic files and databases. We have addressed this problem in a university hospital setting by developing the Tampere Research Archival System (TARAS), an extensive data warehouse for research purposes. This dynamic system includes numerous integrated and pseudonymized imaging studies and clinical data. In a pilot study on asthma patients, we tested and improved the functionality of the data archival system. TARAS is feasible to use in retrieving, analyzing, and processing both image and non-image data. In this paper, we present a detailed workflow of the implementation process of the data warehouse, paying special attention to administrative, ethical, practical, and data security concerns. The establishment of TARAS will enhance and accelerate research practice at Tampere University Hospital, while also improving the safety of patient information as well as the prospects for national and international research collaboration. We hope that much can be learned from our experience of planning, designing, and implementing a research data warehouse combining imaging studies and medical records in a university hospital.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
The advent of the era of electronic Picture Archiving and Communication System (PACS) in the last two decades enabled not only improved diagnostic imaging services, but also the development of multiple useful radiological imaging databases [1–6]. PACS was originally developed for radiology services to replace film-based media with electronically captured medical images, but its use has been extended to other clinical image services as well as collaborations outside the catchment area of a single hospital [7]. Bringing radiology into the electronic era has also come with challenges. The digitization of radiological images has necessitated that hospitals maintain and manage both massive databases of images and a burgeoning IT infrastructure [8].
Clinical databases form an important part of medical research and basic clinical practice. Usually, these databases have been structured into unconnected “silos” of single-purpose data, rather than being combined into a single archive that yields an integrated view of patient data from disparate sources. For example, clinical records and imaging data often exist as separate electronic archives. For clinical research and clinical practice, a joint and dynamic archive would save time, give researchers a full picture of the patient’s condition, and help to find previously unknown associations. Some initiatives have been taken to develop information systems that can integrate biomedical data from multiple laboratories and hospitals. These include the American College of Radiology Imaging Network for cancer studies [9]; the Medical Imaging Resource Center (MIRC) of the Radiological Society of North America, a central repository for clinical trials, research data, and materials [10, 11]; and a Neuroinformatics Database System that supports both prospective and retrospective neuroscience studies [2].
Increases in the amount of electronic patient data [12] highlight the importance of careful data structuring and management. Recent healthcare policies and regulations have affected how patient data is handled in both clinical and research settings. Moreover, the regulations protecting health information have become significantly tighter [13]. Thus, new procedures are needed to facilitate research and ensure data integrity, while also protecting the confidentiality of patient information. Creating an electronic archive involves high costs for purchase and system integration, infrastructure upgrades, risk management, user authorization, engagement of staff, and co-operation between the multiple disciplines within hospitals. Cultural norms for accepting changes likely to bring the hospital to the forefront of high-quality medical research will be needed to accomplish a working system, such as in the adaptation of normal workflow to accommodate PACS [14]. Some initiatives to merge PACS and electronic records have already taken place in patient care settings; this has proven to be a formidable task requiring significant time and attention from hospital CIOs and radiology department managers [15].
This study reviews the authors’ experience in designing and implementing a Research Archival System at Tampere University Hospital, Finland. The Tampere Research Archival System (TARAS) includes two interlinked components: (1) Research PACS, which facilitates clinical imaging research while maintaining the confidentiality and integrity of patient data, and (2) the Clinical Research Data Archival System (CRDAS), which integrates clinical patient data from different information modalities. TARAS is new and separate information system built on top of the existing infrastructure that stores and manages radiological images, processed data from the images (such as lesion volumes), and links with patient records from other medical branches. Thus, the researcher can retrieve all relevant patient information across different systems of data seamlessly, without jeopardizing patient data integrity, normal clinical practice, or data security.
Behind the TARAS initiative of Tampere University Hospital is a greater collaborative goal on both a national and international scale. The aim is to develop a functional setup for the exchange of patient data between research centers and to build a Finnish integrated research center. The realization of this goal would improve access to structured and protected patient data and enhance the feasibility of collaborations between different research groups in Finland and at an international level. This can be achieved by constructing, within the different research centers, separate archives that are consolidated nationally by coherent pseudonymization processes. Pseudonymization is the procedure through which all person-related data are replaced by unique identifiers, allowing the tracking data back to its origins. In anonymization, all person-related data that could allow backtracking is nullified. In our research enterprise, we use pseudonymization, which enables follow-up studies while also maintaining a high level of patient data security.
In this paper, we test the new Research Archival System in a retrospective pilot study. This pilot study addresses patients with asthma as well as the reanalyzed radiology findings of thorax and paranasal sinuses, additional medical history, occupational and habitation information, exposure history to environmental factors, laboratory test, and spirometry results. Regarding gathering and processing data, retrospective studies are challenging because they often include inconsistent diagnostic protocols. However, we are keen to exploit the power of TARAS in conducting a retrospective study with multidisciplinary information.
The five main aims of the initiative are described in Table 1. After assessing the TARAS initiative in a pilot study, we will pseudonymously store numerous radiological images produced at Tampere University Hospital for many different research projects. TARAS is an important step toward combining the radiological images and data contained in the distributed information systems of today’s hospital environment into one research archive. We hope that much can be learnt from the experience of planning, designing, and implementing the combination of Research PACS and CRDAS within a university hospital setting.
Methods
This study describes the fusion of a PACS server (neaPACS®) with a HIS server (CRDAS) and their implementation in a large university hospital organization. To test the combination PACS/HIS software, a pilot study retrospectively examined 120 patients with asthma. Institutional review board permission was granted for the pilot study. The following sections provide a description of the research enterprise workflow, the construction of Research PACS, CRDAS and their fusion, and finally the implementation of the ensemble.
Workflow of the Research Archival System
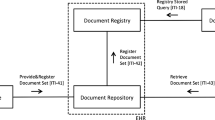
The Medical Imaging Centre and the Science Center of Pirkanmaa Hospital District divided responsibilities to construct, develop, and facilitate the practical use of TARAS. The Science Center is the department responsible for managing scientific studies. The Medical Imaging Centre defines the database variables and transfers images from the clinical PACS to the Research PACS. The Science Center administers the study databases within the CRDAS, the pseudonymization code register, and the Research PACS, as well as data transfer from the external sources. This process is depicted in Fig. 1, which shows the pathway from the request by a researcher to use TARAS to the new research project database and the linked images in the Research PACS. Figure 1 also shows how the system integrates into the organization.
Workflow of the research enterprise. A principal investigator from any clinical department defines, collects, and delivers research information to the main user of CRDAS (Clinical Research Data Archival System) (1), who establishes a new database on the basis of the material received (2). The CRDAS main user designs and carries out the transfer of patient information from other hospital information systems with the respective system administrator (3–4). The main user notifies the principal investigator of the new research project database’s readiness to receive image links (5). The principal investigator delivers the information on the patient images to the imaging coordinator (6). The imaging coordinator conducts the image transfer from clinical PACS to Research PACS through the automated pseudonymization process (7–10). The resulting pseudonymization list is moved to TARAS’s pseudonymization code register (11). The patients’ social security numbers are replaced with pseudonymization codes (12). The main user creates usernames for the defined researcher group and informs them that the new research project database is ready to use (13). When researchers open patient information records, they find a link to the related images in each record and can view the images in a separate image viewer (under the control of Medical Imaging Centre) (14), retrieve the images from the Research PACS (15), and make the image interpretation in the CRDAS database (16)
Construction of Research PACS
The research image archive used to construct Research PACS, neaPACS®, is a standard-conforming Digital Imaging and Communications in Medicine standard (DICOM) PACS augmented with automatic pseudonymization capabilities. The image archive provides standard DICOM PACS services such as storing, querying, image and other DICOM data retrieval, as well as a WWW administrative interface and a viewing application (neaView® radiology) for diagnostic radiological use and administrative purposes.
Each image sent to the archive is automatically pseudonymized according to configurable rules. The identity of the DICOM entity sending the images determines which rule to use. The pseudonymization (or lack thereof) of each DICOM field can be configured separately. Information that could be used to identify the patient or organization can be replaced with generated values that bear no relation to the original values. Certain information of limited sensitivity, e.g., patient’s age, sex, or date of imaging study, can be retained if research purposes deem it necessary. All vendor-specific information is removed because it may contain personal information.
Most of the information replaced with pseudonymized values is permanently discarded; the exception is the single unique identifier for each patient, imaging study, series, and image. The original and corresponding pseudonymized identifiers are stored separately from the pseudonymized images and only used internally by the pseudonymization process to ensure that subsequent data on the same patient, study, or series will be pseudonymized with the same patient, study, or series information respectively, as with previous images. Thus, the hierarchical relationship of data received is maintained after pseudonymization and allows the conduction of longitudinal studies.
In our study, each patient’s social security number was encoded in the Research PACS database by a pseudonymization code combining the encoding day and a running number within the day. If that social security number already exists in the database, the corresponding pseudonymization code is used and no new code is necessary. The following describes a typical configuration of which fields are pseudonymized and how. The PACS software places no other limits on which fields are pseudonymized except that Patient ID and Study, Series, and SOP Instance UIDs must always be assigned new values. In practice, several other values should always be pseudonymized or cleared as well to prevent identifying information from being revealed.
A sample of fields that are assigned new values are listed in Table 2. Fields that are replaced with date or time of pseudonymization are for example birth date of the patient, study date, and study time. The same value is used for study/series level fields in all instances in the same study/series, respectively, as required by DICOM. Fields that are cleared (replaced with an empty value) include referring physician, station, and institution details etc. In addition, all private DICOM fields (odd group number, any element number) are completely removed from the pseudonymized dataset.
Only pseudonymized information is visible to the users and administrators of the archive, regardless of whether the archive is accessed through standard DICOM interfaces (e.g., a researcher’s workstation), the viewer application or the administrative WWW interface. Original information is not displayed to users, and with the exception of the unique identifiers mentioned above, not stored in the image archive. However, it should be noted that automatic pseudonymization is not sufficient to guarantee patient confidentiality; for example, images sent to the archive may contain identifying information such as textual annotations or facial photographs, for which automatic detection and removal are not feasible or reliable. Hence, human involvement is required when selecting images for pseudonymization.
In the Research PACS pilot study, the image data of 120 patients, including most often multiple X-ray and CT imaging studies per patient, were transferred to the Research PACS archive. The transfer was executed with a normal PACS client. The Research PACS archive received image data sent by the clinical PACS client. The transfer procedure can be controlled either manually—as was the case of the pilot study—or automated. If automated, images are sent to the Research PACS database directly from the scanner device in the scanning phase, based on the patient’s social security number or other parameters, if the patient consented to the transfer.
After transfer and pseudonymization are complete, the radiological images can be viewed using the neaView® interface (Fig. 2), which includes several basic image processing tools needed to clinically analyze the images.
Construction of the Clinical Research Data Archival System
Pirkanmaa Hospital District’s Science Center maintains an information system for health research data called the Clinical Research Data Archival System. It is based on the Sapphire LIMS® information system and is customized for scientific database purposes. CRDAS uses Oracle® as a database solution and Business Objects® as a reporting tool. CRDAS has a web-based user interface; access to the system is restricted to the users of the Pirkanmaa Hospital District Intranet. The head of the Science Center grants user rights and roles based on principal investigator’s applications. The main user of CRDAS configures user details and roles. Roles vary from registered users with full access to the data (typing rights, etc.) to view-only access. Only people with user rights can access a specific database. User rights can be modified and expanded after the new research database has been created; for example, in cases of new co-operation projects between different research groups.
The Pirkanmaa Hospital District has many different information systems for patient health care; however, systems do not usually communicate with each other and information is difficult to retrieve from these systems. CRDAS can be configured to have many structural databases according to the researcher’s needs. Data can be stored in the CRDAS by automatic update, manual data submission, or updates from specific files, such as comma separated value files. CRDAS has configurable user rights, database variables, and a graphic interface. Patient data can be normal, pseudonymized, or completely anonymized data. In the pilot study, we used pseudonymized data, which enables us to do follow-up studies. However, anonymization could also be applied in studies using data from deceased patients.
The most recent personal data on a patient can be obtained from the personal data register of Tampere University Hospital via HL7 connection. Most of the patient’s clinical information is obtained from the Information Service of the Hospital, which gathers patient data from various sources and provides sampling options. The Information Service of the Hospital uses an information system called SAS®, which researchers cannot use. Thus, CRDAS is needed to provide greater data access and to serve, metaphorically, as a window with the required information security properties for this large data warehouse.
In the pilot study, the data was collected from patient health records and from radiological image analysis. When collating data from different sources, social security numbers were used to identify the data of a single patient; however, the complete database only used a pseudonymization code as a personal identification. Pseudonymization (Fig. 3) provides a chance to have an open database for different research groups with different research interests and facilitates access to permission rights. As the data is pseudonymized, patient information can be updated in the future. The head of the Science Center controls the pseudonymization codes and can authorize possible decoding.
The principal investigator first lists all data variables needed for the study and then delivers this information to the main user of CRDAS. The main user creates a new database for the study and configures its variables. When the new database is ready for data input, patient data is transferred via a manual input and data transfer. One of the main goals of the TARAS project was to link pseudonymized radiological images to CRDAS so that analysis results of the images can be saved directly in the database.
CRDAS has an easy-to-use user interface and is shown in Fig. 4. The database configuration determines how data is entered. Options include typing or selecting from a list, a data collection, or a calendar. New users may have different options available depending on what role they have been assigned. Patient queries can be done, for example, using the pseudonymization code as an identifier.
Linking Research PACS and Clinical Research Data Archival System
The newly created pairs of social security number and pseudonymization code in the Research PACS were transferred into the CRDAS database in order to enable the combination of patients’ electronic medical records and patients’ images for scientific purposes. However, a TARAS user is not able to see any identifying information. Instead, he or she is able to see patient images and the information saved to the CRDAS (e.g., age, profession, laboratory results, etc.) depending on the specific needs of the study.
We use a straightforward solution in which the user searches images in the neaView® interface (Fig. 2) by entering a pseudonymization code, study time, or image modality issued by CRDAS. However, we plan to create a direct link from the CRDAS user interface to the neaView® interface in the near future.
Implementation of the Research PACS and Clinical Research Data Archival System
Before the final implementation of Research PACS and CRDAS, Dr. Prasun Dastidar, M.D., Ph.D., experienced radiologist, and Ms. Tiina Rajala, M.Sc., Bachelor of Medicine, performed a dummy run using TARAS on a personal computer Osborne Mini 945 workstation with 1 GB memory, 3 GHz Intel Pentium 4 processor, Windows XP operating system (Service Pack 2), and a 19-in. monitor with 1,280 × 1,024 pixel resolution. The findings are presented in the Results and Discussion.
Results and Discussion
Project Timeline and Budget
The TARAS initiative and pilot study represent a step towards a separate large-scale interdisciplinary imaging research database in university hospitals. An initiative of this scale requires a lot of planning, funding, and implementation. Project planning began in January 2008 and the first phase ended with the completion of the pilot study and the testing of TARAS in late 2009. The clinical results of the pilot study will be described in a future publication and thus are not addressed here.
The initial planning of TARAS took approximately 1 year, starting in September 2007 and ending in late September 2008; at this point, the pilot study began. The main personnel involved in software development included three physicians and two engineers, who performed this study as a part-time job. Other personnel involved and interviewed during this study included the department heads of diagnostic radiology and the Science Center (responsible for scientific study management). In addition, four clinicians from the departments of industrial medicine, otorhinolaryngology, and pulmonary and allergic medicine planned the pilot study. Altogether, this project involved 7 months of active research, which made up roughly 36% of the planned budget. The person workload was approximately 77 months and the software costs made up about 47% of the budget. The remaining 17% of the research budget was reserved for software development.
Regarding the amount of time needed for the pilot study: defining and carefully reviewing the data matrix took over a month; patient selection was done within a few weeks; the analysis of the radiological images was done over the course of 2 months; patient data was manually collected for 2 months (it was not in electronic format; however, it is sometimes possible to obtain most of the information directly from the hospital information systems, which accelerates data collection). Additionally, register creation and data configuration took a couple of days, and the manual patient images transfer from the operating PACS into the Research PACS lasted for 4 h. The data matrix definition is probably the most important step and may be the most time-consuming phase in future projects. Data transfer planning must be carefully considered. The other tasks needed to enable data interpretation and research normally last between a couple of weeks to a few months.
Project Achievements
The aims of this project were accomplished successfully. We purchased and set up new Research PACS software (neaPACS®) solely for scientific purposes inside the Medical Imaging Centre under the control of the Science Center of the Pirkanmaa Hospital District. Research PACS was combined with a fairly new clinical records database for research purposes, CRDAS, administered under the Science Center. We named this combination Tampere Research Archival System and tested it in a retrospective pilot study. The components of TARAS worked efficiently and without major problems both alone and together. Specifically, we observed the following in the dummy run:
-
1.
CRDAS: All the necessary clinical, laboratory, and radiological findings could be retrieved in seconds. All the necessary data could be easily found with thumbnail icons. The two trial investigators found that CRDAS was as fast as the existing clinical HIS employed at Tampere University Hospital.
-
2.
The Research PACS server (neaPACS®): User interface tools were well suited for Tampere University Hospital clinicians to analyze radiological images and were easy to adopt. The picture quality and existing post-processing tools were satisfactory. Separate programs can be developed for more detailed analysis of the images (e.g., tissue recognition).
We have compared TARAS to other systems described below. In digital teaching files authoring environment project CASIMAGE [16], a database providing textual fields and annotations related to the patient images was integrated with a PACS server, but did not include other patient information. TARAS also includes well-structured and processed data with hundreds of user-defined parameters that can be retrieved from the original patient archives without compromising security and confidentiality. This enables reliable statistical comparisons within and between patient groups. In another system designed for radiology education with electronic teaching files, SN.MIRC, one has considered patient privacy and data integrity as issues that need to be addressed carefully and concluded that an effort towards a holistic approach should be taken [17]. TARAS fulfills requirements of both privacy and integrity, and can be seen as a concept which greatly improves the research efficiency. Note that because all images are stored in their original format in Research PACS (which currently has 2.8 TB of storage space), there is no loss of image data unlike studies that rely on JPEG images. RADICS—a vendor-neutral digital teaching file system—emphasizes the importance of minimizing clinical workflow disruption when image studies are copied into a research archive [18]; however, TARAS enables fast and reliable export of patient data during a clinical study. The ease of gathering a vast amount of data and the high level of integration when compared to many other research databases are key factors for successful implementation of medical research projects. Based on these comparisons, we believe that the TARAS server is at least as comprehensive as the systems mentioned earlier.
Data Management
Data protection is also an important topic in the storage of vast quantities of data. The fact that both Research PACS and CRDAS are not connected to the Internet reduces the risk of someone hacking into the system and compromising data confidentiality. Also, in case of the system crashing, information lost from the Research PACS or CRDAS can be restored within the last month. In the future, unless needed for interfacing to external systems (CRDAS, permission management), the original unique identifiers do not need to be stored in the archive. It would be sufficient to record cryptographically secure one-way hash values (i.e., checksums) of the identifiers. From the hash values, the original values cannot be recovered. Thus, no identifying information could be revealed even if archive security was compromised. These data security, confidentiality, and backup measures guarantee that the system is sufficiently foolproof. The involvement of data security personnel in TARAS’s construction phase was an important factor and helped to avoid conflict situations with increasing demands for patient confidentiality as well as the need for data in medical research.
Situations in which investigators want to combine retrospective studies with prospective patient recruitment can be handled so that data is collected with the patients’ personal code to a temporary database. After data collection is ready, personal information is pseudonymized and it is possible to merge databases. In a case where pseudonymization code has to be opened, it is done by the TARAS main user.
In its present form, Research PACS has the capability to store images on a scale of 105–108, depending on the image type, quality, size, and other factors. Pictures are stored on their own server whose memory can be expanded. The pseudonymization process is relatively lightweight and adds negligible overhead to image reception. The amount of image data that can be stored and retrieved is for practical purposes limited only by available disk space, which can be expanded as required. These things indicate that if a system works with smaller amount of data, it is easily scalable. Regarding CRDAS’s use as a silo for electronic patient records, its memory capacity is limitless. CRDAS uses Oracle as a database solution and it can be updated to answer future demands. On general terms we see no substantial additions to costs at technical side in near future. Additional costs will arise with the running salary of one employee.
The quality control of TARAS has been carefully considered to enable integrated, seamless, and accurate information flow in the system infrastructure. In the system implementation phase, a detailed data cross-validation and review processes have been performed to fulfill the requirements of robust architecture. Data integrity and correctness are also tested frequently during the research project to reveal possible inconsistencies. Validation of the pseudonymization was performed by verifying that no identifying information was visible in neaView®. Furthermore, the DICOM headers of a random sample of pseudonymized images were manually examined and the absence of patient-identifying information verified. Automatic validation of the pseudonymization has been considered as a possible future enhancement.
Thus, the data warehouse system for research purposes was successfully implemented and operational in the pilot study; however, improvements can still take place. Future plans include more seamless integration of the image archive with the CRDAS, whereby archived images could be viewed using a link on the CRDAS patient view page. In addition, the Research PACS includes only native X-ray and CT images, but different image modalities will be added during forthcoming research projects. The addition of different imaging modalities will be a straightforward task because Research PACS is able to receive several types of medical images. Moreover, the electronic availability of patient records in a structured format is, at the moment, a limiting factor in the automatic collection of patient data into the CRDAS. For example, laboratory tests and diagnoses can be transferred automatically into the CRDAS, but many patient records are not structured so that they could be saved into CRDAS in a searchable format.
A separate TARAS workstation is being designed and built within the Medical Imaging Centre under the administration of the Science Center for everyday use. This workstation includes two monitors, so that both Research PACS and CRDAS user interfaces can be opened at the same time. The TARAS workstation also facilitates a more detailed toolbox for radiological image analysis because there will be several applications that the user may use for research purposes. These tools include 3D volumetric and segmentation applications as well as a program for the texture analysis of medical images.
Lessons Learned
The main lesson learned from the planning and implementation of a research archive was that the involvement of experts in different clinical and technical areas is important to achieve a functional Research Archival System. Regular meetings with personnel from these different areas are important to understand and create a common language. In the pilot study, it became more than obvious that preliminary detailed clinical data and different variables for each patient to be saved in the CRDAS must be precisely thought, especially concerning the main clinical aims. The data collected in TARAS is a valuable silo for research projects in the future.
The projects incorporated into TARAS must have a statement from the ethics committee and must be carried out according to the Declaration of Helsinki. The study subjects must also give written informed consent to the work. The system is built so that neither patient privacy nor hospital clinical practice is compromised in any way during the research protocol. Only a designated individual with the relevant ethics training will have access to the pseudonymization key and this person will not be directly involved in the research project. The head of the Science Center is responsible for deciding who may access and supervise the specific project databases. The principal investigator will be responsible and apply for participant user rights from the head of the Science Center. Members of a research project will only have access to the research project database to which they have user rights. Only the principal investigator may export the data matrix from CRDAS with authorization from the head of the Science Center. The question remains as to whether a special ethics committee is needed to oversee the various research projects in TARAS.
Our project has been a successful transition from old-fashioned and separate data archives toward an integrated, fast, reliable, and easy-to-use system. Although the pilot study has proven TARAS’s viability, combination development will further enhance its capacity and enable user-friendly utilization of research information.
Future Trends and Conclusions
Future Trends
The process of achieving this research archive has taught us many pros and cons that need to be addressed here. We believe that this research archive, which is currently the only one of its kind in Finland, needs further diversification into a national research archive. The Social Insurance Institution of Finland is currently engaged in integrating all existing local clinical PACS/HIS systems into one national clinical PACS/HIS system for use by the year 2012. This integration will be of enormous help to all hospitals in Finland because transferring clinical, laboratory, and imaging data between different health centers, referral hospitals, central hospitals, and university hospitals will become a simple efficient reality. We believe that if we could create a similar kind of research archive with national integration, it would benefit those interested scientific study groups and institutions working at national level and, eventually, also at the international level.
Of course, such an informatics system requires monetary resources. When developing such a system, one should keep in mind the different obstacles such as the cultural, religious, medico-legal, and ethical issues in a given country and respect these aspects in the final integration. Healthcare resources prove to be a major obstacle for this kind of archive; hence, active involvement of all the major hospital administrators is an important work goal. These administrators should learn to appreciate the value of such an archive in research and development, as well as daily clinical work. As the technology advances, one must constantly update the software and monitors every year to keep the archive up to par and also increase the memory storage capacity for bigger and more challenging research enterprises. In addition, the integration of national data archives (e.g., FINJEM, a Finnish job exposure matrix already incorporated into the CRDAS [19]) should be a priority.
Conclusion
In conclusion, we introduced a large-scale system that has removed the traditional barriers between clinical images and other patient records, thereby providing a single source of information for multiple areas of medical research. This clinical information is pseudonymized, guaranteeing patient confidentiality and ease for research work as well as enabling follow-up studies. Notably, the information system is separate from the actual hospital data archives, thereby ensuring data integrity and availability for routine clinical work.
References
Rosset A, Ratib O, Geissbuhler A, Vallée JP: Integration of a multimedia teaching and reference database in a PACS environment. Radiographics 22:1567–77, 2002
Wong ST, Hoo Jr, KS, Cao X, Tjandra D, Fu JC, Dillon WP: A neuroinformatics database system for disease-oriented neuroimaging research. Acad Radiol 11:345–58, 2004
Sasso G, Marsiglia HR, Pigatto F, Basilicata A, Gargiulo M, Abate AF, Nappi M, Pulley J, Sasso FS: A visual query-by-example image database for chest CT images: potential role as a decision and educational support tool for radiologists. J Digit Imaging 18:78–84, 2005
Marcus DS, Archie KA, Olsen TR, Ramaratnam M: The open-source neuroimaging research enterprise. J Digit Imaging 20:130–8, 2007
Yang GL, Tan YF, Loh SC, Lim CC: Neuroradiology imaging database: using picture archive and communication systems for brain tumour research. Singapore Med J 48:342–6, 2007
Ng CK, White P, McKay JC: Development of a web database portfolio system with PACS connectivity for undergraduate health education and continuing professional development. Comput Methods Programs Biomed 94:26–38, 2009
Huang HK: Enterprise PACS and image distribution. Comput Med Imaging Graph 27:241–53, 2003
Channin DS, Bowers G, Nagy P: Should radiology IT be owned by the chief information officer? J Digit Imaging 22:218–21, 2009
Hillman BJ: The American College of Radiology Imaging Network (ACRIN): research educational opportunities for academic radiology. Acad Radiol 9:561–2, 2002
Gentili A, Chung CB, Hughes T: Informatics in radiology: use of the MIRC DICOM service for clinical trials to automatically create teaching file cases from PACS. Radiographics 27:269–75, 2007
Tellis WM, Andriole KP: Implementing a MIRC query interface for a database driven teaching file. J Digit Imaging 16:180–4, 2003
Haux R: Health information systems—past, present, future. Int J Med Inform 75:268–81, 2006
Bland PH, Laderach GE, Meyer CR: A web-based interface for communication of data between the clinical and research environments without revealing identifying information. Acad Radiol 14:757–64, 2007
Paré G, Trudel MC: Knowledge barriers to PACS adoption and implementation in hospitals. Int J Med Inform 76:22–33, 2007
Briggs B: Melding PACS and electronic records. Health Data Manag 13:28–30, 2005
Rosset A, Muller H, Martins M, Dfouni N, Vallée JP, Ratib O: Casimage project: a digital teaching files authoring environment. J Thorac Imaging 19:103–8, 2004
Yang GL, Lim CC: Singapore national medical image resource centre (SN.MIRC): a world wide web resource for radiology education. Ann Acad Med Singapore 35:558–63, 2006
Kamauu A, DuWall S, Robison R, Liimatta A, Wiggins III, R, Avrin D: Vendor-neutral case input into a server-based digital teaching file system. RadioGraphics 26(6):1877–1885, 2006
Kauppinen T, Toikkanen J, Pukkala E: From cross-tabulations to multipurpose exposure information systems: a new job-exposure matrix. Am J Ind Med 33(4):409–17, 1998
Conflicts of interest
The authors have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Rajala, T., Savio, S., Penttinen, J. et al. Development of a Research Dedicated Archival System (TARAS) in a University Hospital. J Digit Imaging 24, 864–873 (2011). https://doi.org/10.1007/s10278-010-9350-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-010-9350-1