Abstract
Periodontitis is a multifactorial disease associated with genetic and environmental factors. Single-nucleotide polymorphisms (SNPs) are associated with susceptibility to common diseases such as diabetes and periodontitis. Although the oral cavity is exposed to various organisms, the conditions are well controlled by innate and acquired immune systems. Antimicrobial peptides (AMPs) play an important role in the innate immune system; however, the association of AMP-SNPs with periodontitis has not been fully elucidated. This study investigated the relationship between AMP-SNPs and periodontitis in Japanese. One hundred and five Japanese subjects were recruited, which included patients with aggressive, severe, moderate and mild periodontitis, and age-matched healthy controls. Genomic DNA was isolated from peripheral blood and genotypes of SNPs of β-defensin-1 and lactoferrin genes (DEFB1: rs1799946, rs1800972 and rs11362; and LTF: rs1126478) were investigated using the PCR-Invader assay. Protein level of AMPs in gingival crevicular fluid (GCF) was quantified by ELISA. Case–control studies revealed that the −44 CC genotype of DEFB1 (rs1800972) was associated with periodontitis (OR 2.51), particularly with severe chronic periodontitis (OR 4.15) and with combined severe and moderate chronic periodontitis (OR 4.04). No statistical differences were found in other genotypes. The β-defensin-1 concentrations in GCF were significantly lower in subjects with the −44 CC genotype of DEFB1 than in those without this genotype. No significant differences between GCF concentrations of AMPs and other genotypes were detected. The −44 CC genotype of the β-defensin-1 gene (DEFB1 rs1800972) may be associated with susceptibility to chronic periodontitis in Japanese.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Periodontitis is an inflammatory disease primarily caused by the infection of dental plaque microorganisms, followed by periodontal tissue destruction after a continuous excessive immune response. There are many factors that initiate and progress periodontitis, which include not only microbial factors but also other local and systemic factors such as age, gender, ethnicity, smoking, lifestyle, stress, heredity and systemic diseases. Thus, periodontitis is considered to be a multifactorial disease associated with genetic and environmental factors [1]. Genetic factors can convey susceptibility to periodontitis and recent reports have shown that single-nucleotide polymorphisms (SNPs) are important variations among the genetic factors that underlie the host response to diseases [2, 3]. In general, humans have 2–3 million SNPs, which correspond to 0.1 % of genomic DNA. SNPs in non-coding regions can affect the regulation of gene expression, and SNPs in coding regions can change protein sequences and often give rise to different biological functions. Many basic and clinical studies have shown the relationship between SNPs and susceptibility to periodontitis [2–4]. It has been reported that SNPs of interleukin (IL)-1, IL-6, IL-10, IL-17, tumor necrosis factor (TNF)-α and immunoglobulin G Fc receptors (FcγR) may be associated with the initiation and progression of periodontitis [5–10]. Our collaborating studies reported that vitamin D receptor (VDR) +1056 polymorphisms were related to chronic periodontitis [11] and that α2 integrin +807 polymorphisms were related to drug-induced gingival overgrowth [12].
The oral cavity is continuously exposed to various pathogenic organisms, but oral conditions are well controlled by innate and acquired immune systems. Innate immunity functions in most species, including insects, fungi, plants and mammals, as non-specific host defense at an early stage of microorganism challenge. Antimicrobial peptides (AMPs) play an important role in the innate immune system and more than 800 AMPs have been identified [13, 14]. Many AMPs such as lysozyme, defensin, lactoferrin, histatin, cystatin and calprotectin were found in the human oral cavity, and several reports demonstrated the association between periodontitis and AMP levels in saliva, gingival tissue or gingival crevicular fluid (GCF) [15–17]. We previously reported that the calprotectin level in GCF correlated with gingival index in periodontitis patients [18, 19]. It is conceivable that gene polymorphisms of AMPs cause differences in the innate immune system and confer susceptibility to various infectious diseases. However, there have been few reports concerning the association of AMP-SNPs with periodontitis.
Human β-defensins are small cationic AMPs, and at least four types of β-defensin have been characterized [20]. β-defensin-1 is a prominent molecule of the defensin family that is encoded by the DEFB1 gene [21]. SNPs of the DEFB1 gene are located in coding and non-coding regions, including the 5′-untranslated region (5′-UTR) and 3′-UTR [22, 23]. β-Defensin-1 is expressed in keratinocytes and epithelial cells, and is found in the kidney, female reproductive tract, testis, gingival tissue, small intestine, large intestine, cornea and mammary gland [22, 24]. Lactoferrin, also known as lactotransferrin, is a multifunctional metalloprotein, which is a member of the transferrin family [25]. Human lactoferrin is encoded by the LTF gene and SNPs of the LTF gene have been identified in both coding and non-coding regions [26]. Human lactoferrin is produced by mucosal epithelial cells and widely found in various secretory fluids, such as milk, saliva, tears, sweat and nasal secretions [25]. In addition, human lactoferrin is produced by secondary granules of neutrophils and is released in response to inflammation [27].
In this study, we analyzed the genotype distributions and allele frequencies of three DEFB1 SNPs and one LTF-SNP in Japanese periodontitis patients, and then we quantified the GCF concentrations of these AMPs in the subjects. Since SNPs are generally associated with ethnicity and geography, this study focused on Japanese subjects to determine the relationship between AMP-SNPs and chronic or aggressive periodontitis.
Materials and methods
Subjects and study protocol
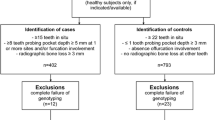
The protocol for this study was reviewed and approved by the ethics committee of human genome and gene analysis at the University of Tokushima (Approval No. H23-7). From subjects who agreed to participate in the study, written informed consent was obtained before undergoing periodontal examinations. Periodontal conditions were diagnosed according to the criteria of the 1999 International Workshop for Classification of Periodontal Diseases and Conditions [28]. The subjects were divided into five groups: chronic periodontitis (CP) including severe CP, moderate CP and mild CP, aggressive periodontitis (AgP) and age-matched healthy controls, on the basis of clinical examinations including probing depth (PD), clinical attachment level (CAL), bleeding on probing (BOP) and alveolar bone loss (BL). The bone loss was assessed at 2 sites of a tooth on a radiograph [29]. Exclusion criteria were the presence of systemic disease (e.g., diabetes mellitus and kidney disease), drug-induced gingival overgrowth, pregnancy, having fewer than 15 teeth and a history of any periodontal therapy within the previous 6 months. Age-matched healthy controls were subjects more than 40 years old in the case of mild CP and under 35 years old in the case of AgP, both having localized PD ≤3 mm and mean BL ≤15 %. The severe CP group consisted of subjects more than 40 years old having localized PD ≥4 mm and mean BL ≥34 %, and the moderate CP group consisted of subjects more than 40 years old having localized PD ≥4 mm and 17 % ≤ mean BL ≤ 28 %. The AgP group consisted of subjects under 35 years old having localized CAL ≥5 mm. One hundred and five Japanese subjects were recruited, including 62 periodontitis (28 with severe CP, 13 with moderate CP and 21 with AgP) and 43 controls (22 for CP and 21 for AgP).
Isolation of genomic DNA and genotype determination
Five milliliters of peripheral blood was obtained from the basilic, cephalic or median cubital vein of each subject. Genomic DNA was isolated from the blood sample and the genotypes of the β-defensin-1 gene (DEFB1) and the lactoferrin gene (LTF) were determined using a PCR-Invader assay by BML Inc. (Saitama, Japan). The PCR-Invader assay was reported previously [30]. We assayed three known SNPs in the 5′-UTR of DEFB1, namely, at positions −52 G/A (rs1799946; a guanine to adenine nucleotide mutation), −44 C/G (rs1800972; a cytosine to guanine nucleotide mutation) and −20 G/A (rs11362; a guanine to adenine nucleotide mutation) from the first AUG codon [31–33]. In addition, we assayed one known SNP of LTF, namely, at position 29 in the N-terminal α-helical region of human lactoferrin (rs1126478; an adenine to guanine nucleotide mutation) [34, 35].
Sampling of gingival crevicular fluid and β-defensin-1 quantification
Gingival crevicular fluid (GCF) samples were collected according to a method described previously [36]. Briefly, before the clinical evaluation including PD and BOP, GCF was collected using Periopaper® (Oraflow Inc., New York, NY) from healthy gingival crevices or periodontal pockets with periodontitis. GCF volume was determined using Periotron 8000® (Harco Electronics, Winnipeg, MB, Canada). The GCF perfused in paper strips was extracted using 100 μl of 10 mM Tris–HCl buffer (pH 7.4) containing 200 μM phenylmethylsulfonyl fluoride. The amounts of β-defensin-1 and lactoferrin in GCF samples were determined using enzyme-linked immunosorbent assay (ELISA) kits (Human BD-1 ELISA Kit, pink-ONE, KOMA BOTEC Inc., Seoul, Korea; Human Lactoferrin ELISA Kit; HyCult Biotechnology b.v., Uden, Netherlands). The samples were diluted 2- or 10-fold with the dilution buffer of each kit and used for the determination of β-defensin-1 and lactoferrin according to the manufacturers’ instructions. After the total amounts of β-defensin-1 and lactoferrin were measured from the standard curve, the β-defensin-1 and lactoferrin concentrations of GCF samples were obtained as pg or ng/μl by adjusting for the GCF volume.
Statistical analyses
The distribution of each SNP genotype was evaluated for Hardy–Weinberg equilibrium. Statistical analyses were performed using JMP® software (SAS Institute Inc., Cary, NC, USA). The differences in genotype distributions and allele frequencies were tested by Chi-square tests. When an expected value in cells was less than five, Fisher’s exact tests were used. Contingency tables (2 × 2) were used for four kinds of comparison: periodontitis vs. controls, severe CP vs. CP control, combined periodontitis (severe and moderate CPs) vs. CP control and AgP vs. AgP control. The strength of the associations was determined using odds ratio (OR) calculation and 95 % confidence intervals (95 % CI). In the ELISA analysis, the relationships between β-defensin-1 concentration in GCF and DEFB1 genotypes and between lactoferrin concentration in GCF and LTF genotype were evaluated using the nonparametric Wilcoxon signed-rank test and Mann–Whitney U test. A p value of <0.05 was considered to be statistically significant.
Results
Demographics
Table 1 provides a summary of the demographic and clinical characteristics of the 105 Japanese subjects, 62 periodontitis and 43 controls. The table also provides information of the five groups, consisting of severe CP, moderate CP, AgP, CP control and AgP control. Periodontitis subjects showed significantly higher values of BL, PPD and BOP than the controls (p < 0.01). The mean ages of those with severe CP, moderate CP and CP control were similar, whereas those with AgP were significantly older than those in the AgP control group (p < 0.05). The severe CP group showed significantly higher values of BL, PPD and BOP than the CP control group. The moderate CP group showed a significantly higher value of BL than the CP control. When the severe and moderate CPs were combined (n = 41), higher values of BL, PPD and BOP were observed in the combined group than in the CP control group (p < 0.01, data not shown). The AgP group showed significantly higher values of BL, PPD and BOP than the AgP control.
SNP analysis of DEFB1 and LTF
We analyzed three SNPs at positions −52 G/A, −44 C/G and −20 G/A in the 5′-UTR of DEFB1 and one SNP at position 29, Lys/Arg, in the N-terminal alpha-helical region of human lactoferrin. The genotype distribution of these SNPs did not show any significant difference from Hardy–Weinberg equilibrium. The genotype distributions and allele frequencies of DEFB1 and LTF polymorphisms are summarized in Tables 2, 3 and 4. The frequency of the −44 CC genotype of DEFB1 was higher in the periodontitis (83.9 %) than in the controls (67.4 %) (Table 2). In the case of the CP groups, the frequency of the −44 CC genotype of DEFB1 was higher in the severe CP group (85.7 %) and the moderate CP group (84.6 %) than in the CP control (59.1 %) and the frequency of the −44 C allele was higher in the severe CP group (92.9 %) than in the CP control (79.5 %) (Table 3). In the case of the AgP group, no difference of −44 CC genotype or the −44 C allele was observed between AgP and its control (Table 4). These results indicate that not only the frequency of the −44 CC genotype of DEFB1 was relatively high in the periodontitis, but also the frequencies of the −44 CC genotype and the −44 C allele of DEFB1 were relatively high in cases of CP. Next, we performed four kinds of comparison: periodontitis vs. controls, severe CP vs. CP control, combined periodontitis (severe and moderate CPs) vs. CP control and AgP vs. AgP control for genotypic/allelic associations of the DEFB1 and LTF polymorphisms. In the −44 C/G polymorphism of DEFB1, statistical significance was observed between the −44 CC genotype and periodontitis (OR 2.510, 95 % CI 1.005–6.263, p < 0.05, Table 2). Furthermore, in the −44 C/G polymorphism of DEFB1, statistical significance was observed between the −44 CC genotype and severe CP (OR 4.154, 95 % CI 1.113–15.304, p < 0.05, Table 3) and between the −44 CC genotype and combined periodontitis (OR 4.038, 95 % CI 1.236–13.186, p < 0.05, Table 3). Statistical significance was also identified between the −44 C allele and severe CP (OR 3.343, 95 % CI 1.003–11.041, p < 0.05, Table 3). There were no significant differences in the genotypic/allelic associations of other DEFB1 and LTF SNPs for severe CP vs. CP control and combined periodontitis vs. CP control (Table 3). On the other hand, there were no genotypic/allelic associations of the DEFB1 and LTF polymorphisms between AgP and AgP control (Table 4).
GCF analysis of β-defensin-1 and lactoferrin
To evaluate the relationship between the genotype and expressions of AMPs in GCF samples, 67 GCF samples from 17 subjects and 71 GCF samples from 16 subjects were used for the determination of β-defensin-1 and lactoferrin, respectively. As shown in Fig. 1a, ELISA analysis revealed that the β-defensin-1 concentrations were significantly lower in the subjects with the −44 CC genotype than in those without the −44 CC genotype, namely, 0.95 ± 0.36 and 1.55 ± 0.63 pg/μl, respectively (p < 0.05). Moreover, the β-defensin-1 concentrations in PD ≤3 mm were significantly lower in the subjects with the −44 CC genotype than in those without it, namely, 1.12 ± 0.43 and 2.20 ± 1.22 pg/μl, respectively (p < 0.05, Fig. 1a). There were no significant differences in PD ≥4 mm between the subjects with the −44 CC genotype and those without it, with values of 0.86 ± 0.54 and 0.81 ± 0.13 pg/μl, respectively (Fig. 1a). When the concentrations of lactoferrin in GCF were evaluated, no significant differences were observed between the subjects with the GG genotype of LTF and those without it, with values of 7.77 ± 2.09 and 8.14 ± 4.85 ng/μl, respectively (Fig. 1b).
Associations between the genotypes and concentrations of antimicrobial peptides in GCF samples. GCF samples were collected using paper strips and the volumes were quantified using a calibrated unit. The contents of β-defensin-1 and lactoferrin in GCF were determined using ELISA kits. β-defensin-1 concentrations of subjects with the CC, CG and GG genotypes of rs1800972 (a) and lactoferrin concentrations of subjects with the GG, AG and AA genotypes of rs1126478 (b) are shown as the mean ± SD. *p < 0.05 vs. control
Discussion
In this study, we investigated the association of four AMP-SNPs with the susceptibility to periodontitis in 105 Japanese subjects. We selected three SNPs in the 5′-UTR of the DEFB1 gene (rs1799946, rs1800972 and rs11362) and one SNP in exon 1 of the LTF gene (rs1126478). These SNPs have been considered to be associated with various infectious diseases. DEFB1 gene polymorphisms were reportedly related to Pseudomonas aeruginosa airway colonization in cystic fibrosis, oral Candida carriage, lepromatous leprosy, HIV infection, Helicobacter pylori-induced gastritis, severe acute pancreatitis and dental caries [31, 33, 37–41]. LTF gene polymorphism was reportedly related to dental caries [42]. In terms of the SNPs in the 5′-UTR of DEFB1, −44 C/G was not associated with early-onset periodontitis [43] and that −20 G/A was not associated with severe CP [32]. In the case of LTF-SNP, Lys/Arg polymorphism was shown to be associated with AgP [34, 35]. In this study, we provided the first evidence that the −44 CC genotype of DEFB1 was associated with periodontitis (OR 2.510), particularly with severe CP and severe/moderate-combined periodontitis (OR 4.154 and 4.038, respectively), whereas there was no association with AgP. This result is similar to a previous report that showed that the −44 C/G polymorphism was not associated with AgP [43].
The present finding, the association of −44 CC genotype with CP, may be linked to the low expression of β-defensin-1 in gingival tissue, because basal levels of β-defensin-1 reflect a protective ability in localized tissues [44, 45]. Our result suggests that the −44 C/G polymorphism may affect β-defensin-1 expression in gingival tissue and enhance the disease susceptibility to CP. It was reported that the constitutive human β-defensin-1 mRNA level was lower in primary gingival keratinocytes associated with the −44 CC genotype than in cells associated with the −44 GG and −44 GC genotypes, and that the −44 G allele was associated with an increase in constitutive antimicrobial activity and expression of β-defensin-1 [46]. Recent study concerning lepromatous leprosy demonstrated that the position −44 was included in the putative NFκB binding site and the variant could change the NFκB binding affinity and thus influenced the regulation of DEFB1 gene expression at the transcription stage [37]. Furthermore, several reports showed the homology score between the region from positions −57 to −15 of DEFB1 and the NFκB binding site, and the value for the −44 C allele was shown to be lower than that for the −44 G allele (homology scores of 64.5 and 69.8 %, respectively), indicating that the −44 C/G polymorphism could contribute to susceptibility to infectious disease [37, 47, 48]. Our finding that the subjects with the −44 CC genotype exhibited low concentrations of β-defensin-1 in GCF might be attributable to the −44 C allele that being involved in a decrease in NFκB binding affinity. Since the association between −44 C/G polymorphism and NFκB binding affinity is not completely understood, further study including gel mobility shift assay is necessary to clarify the mechanism involved.
The present finding that the β-defensin-1 concentrations in localized PD ≤3 mm, unlike in PD ≥4 mm, were low in the subjects with the −44 CC genotype suggests that the −44 CC genotype may be related to the onset of CP. However, the subjects with the −44 CC genotype did not always suffer from CP, suggesting that periodontitis is a multifactorial disease associated with not only genetic factors but also environmental ones. It is generally known that β-defensins are expressed in mucosal epithelial cells and keratinocytes to protect oral mucosal surfaces. β-defensin-1 was shown to be expressed constitutively in epithelial tissue, but β-defensin-2, -3 and -4 were found to be up-regulated by proinflammatory cytokines and microorganisms [16, 21]. β-defensin-1, -2 and -3 were expressed in both inflamed and non-inflamed gingival tissues, whereas differential expressions were observed among tissues from healthy subjects, and cases of gingivitis and periodontitis [49–51]. The level of β-defensin-1 mRNA expression was low in cases of gingivitis and AgP, but high in CP; conversely, the level of β-defensin-2 mRNA expression was high in AgP but low in CP [52]. From these findings, our result may reflect that β-defensin-1 plays a role in defense of the gingival mucosal surface.
In this study, we demonstrated that the −52 G/A and −20 G/A polymorphisms of β-defensin-1 were not related to periodontitis including CP and AgP. This result is partially consistent with a previous report [32]. Although previous reports showed that the Lys/Arg polymorphism of lactoferrin was associated with AgP [34, 35], our result showed that there were no significant differences in the associations of Lys/Arg polymorphism with periodontitis including CP and AgP. In addition, the concentrations of lactoferrin in GCF did not show any differences between the subjects with and without the GG genotype. It has been reported that Lys/Arg polymorphism of lactoferrin might change the protein sequence and give rise to reduce antimicrobial activity of lactoferrin [34]. However, our results showed that the Lys/Arg polymorphism was not related to CP and AgP in Japanese patients. We assume that these results might be due to ethnicity and geography because some reports have indicated that a Thr/Ala polymorphism at position 11 resulting from an A/G transition was associated with AgP in African–Americans but not in Caucasians [53].
Taken together, we conclude that the −44 CC genotype of DEFB1 is associated with the susceptibility to periodontitis particularly CP in Japanese and that −44 C/G polymorphism may be a predictor of CP.
Conclusion
The −44 CC genotype of the β-defensin-1 gene (DEFB1 rs1800972) may be associated with susceptibility to chronic periodontitis in Japanese.
References
Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:138–53.
Kinane DF, Shiba H, Hart TC. The genetic basis of periodontitis. Periodontol 2000. 2005;39:91–117.
Yoshie H, Kobayashi T, Tai H, Galicia JC. The role of genetic polymorphisms in periodontitis. Periodontol 2000. 2007;43:102–32.
Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004;317:887–92.
Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, Wilson TG Jr, Higginbottom FL, Duff GW. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–7.
Trevilatto PC, Scarel-Caminaga RM, de Brito RB, de Souza AP Jr, Line SR. Polymorphism at position −174 of IL-6 gene is associated with susceptibility to chronic periodontitis in a Caucasian Brazilian population. J Clin Periodontol. 2003;30:438–42.
Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB, Camargo LE, Line SR. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. J Clin Periodontol. 2004;31:443–8.
Kadkhodazadeh M, Baghani Z, Ebadian AR, Youssefi N, Mehdizadeh AR, Azimi N. IL-17 gene polymorphism is associated with chronic periodontitis and peri-implantitis in Iranian patients: a cross-sectional study. Immunol Invest. 2013;42:156–63.
Soga Y, Nishimura F, Ohyama H, Maeda H, Takashiba S, Murayama Y. Tumor necrosis factor-α gene (TNF-α) −1031/−863, −857 single-nucleotide polymorphisms (SNPs) are associated with severe adult periodontitis in Japanese. J Clin Periodontol. 2003;30:524–31.
Kobayashi T, Yamamoto K, Sugita N, van der Pol WL, Yasuda K, Kaneko S, van de Winkel JG, Yoshie H. The Fc γ receptor genotype as a severity factor for chronic periodontitis in Japanese patients. J Periodontol. 2001;72:1324–31.
Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009;88:1137–41.
Ogino M, Kido J, Bando M, Hayashi N, Wada C, Nagata T, Nishimura F, Soga Y, Takashiba S, Kubota T, Itagaki M, Shimada Y, Tai H, Yoshie H, Yamazaki N, Shinohara Y, Kataoka M. Alpha 2 integrin +807 polymorphism in drug-induced gingival overgrowth. J Dent Res. 2005;84:1183–6.
Kinane DF, Demuth DR, Gorr SU, Hajishengallis GN, Martin MH. Human variability in innate immunity. Periodontol 2000. 2007;45:14–34.
Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–41.
Jentsch H, Sievert Y, Göcke R. Lactoferrin and other markers from gingival crevicular fluid and saliva before and after periodontal treatment. J Clin Periodontol. 2004;31:511–4.
Lu Q, Jin L, Darveau RP, Samaranayake LP. Expression of human β-defensins-1 and -2 peptides in unresolved chronic periodontitis. J Periodontal Res. 2004;39:221–7.
Tsai CC, Kao CC, Chen CC. Gingival crevicular fluid lactoferrin levels in adult periodontitis patients. Aust Dent J. 1998;43:40–4.
Nakamura T, Kido J, Kido R, Ohishi K, Yamauchi N, Kataoka M, Nagata T. The association of calprotectin level in gingival crevicular fluid with gingival index and the activities of collagenase and aspartate aminotransferase in adult periodontitis patients. J Periodontol. 2000;71:361–7.
Kido J, Nakamura T, Kido R, Ohishi K, Yamauchi N, Kataoka M, Nagata T. Calprotectin in gingival crevicular fluid correlates with clinical and biochemical markers of periodontal disease. J Clin Periodontol. 1999;26:653–7.
Diamond G, Ryan L. β-Defensins: what are they really doing in the oral cavity? Oral Dis. 2011;17:628–35.
Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human β-defensins. Cell Mol Life Sci. 2006;63:1294–313.
Prado-Montes de Oca E. Human β-defensin 1: a restless warrior against allergies, infections and cancer. Int J Biochem Cell Biol. 2010;42:800–4.
Dörk T, Stuhrmann M. Polymorphisms of the human β-defensin-1 gene. Mol Cell Probes. 1998;12:171–3.
Dale BA, Krisanaprakornkit S. Defensin antimicrobial peptides in the oral cavity. J Oral Pathol Med. 2001;30:321–7.
García-Montoya IA, Cendón TS, Arévalo-Gallegos S, Rascón-Cruz Q. Lactoferrin a multiple bioactive protein: an overview. Biochim Biophys Acta. 2012;1820:226–36.
Teng CT, Gladwell W. Single nucleotide polymorphisms (SNPs) in human lactoferrin gene. Biochem Cell Biol. 2006;84:381–4.
Legrand D, Elass E, Carpentier M, Mazurier J. Lactoferrin: a modulator of immune and inflammatory responses. Cell Mol Life Sci. 2005;62:2549–59.
Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6.
Schei O, Waerhaug J, Lovdal A, Arno A. Alveolar bone loss as related to oral hygiene and age. J Periodontol. 1959;30:7–16.
Mein CA, Barratt BJ, Dunn MG, Siegmund T, Smith AN, Esposito L, Nutland S, Stevens HE, Wilson AJ, Phillips MS, Jarvis N, Law S, de Arruda M, Todd JA. Evaluation of single nucleotide polymorphism typing with invader on PCR amplicons and its automation. Genome Res. 2000;10:330–43.
Tesse R, Cardinale F, Santostasi T, Polizzi A, Manca A, Mappa L, Iacoviello G, De Robertis F, Logrillo VP, Armenio L. Association of β-defensin-1 gene polymorphisms with Pseudomonas aeruginosa airway colonization in cystic fibrosis. Genes Immun. 2008;9:57–60.
Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006;77:426–36.
Jurevic RJ, Bai M, Chadwick RB, White TC, Dale BA. Single-nucleotide polymorphisms (SNPs) in human β-defensin 1: high-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J Clin Microbiol. 2003;41:90–6.
Velliyagounder K, Kaplan JB, Furgang D, Legarda D, Diamond G, Parkin RE, Fine DH. One of two human lactoferrin variants exhibits increased antibacterial and transcriptional activation activities and is associated with localized juvenile periodontitis. Infect Immun. 2003;71:6141–7.
Wu YM, Juo SH, Ho YP, Ho KY, Yang YH, Tsai CC. Association between lactoferrin gene polymorphisms and aggressive periodontitis among Taiwanese patients. J Periodontal Res. 2009;44:418–24.
Kido J, Nakamura T, Asahara Y, Sawa T, Kohri K, Nagata T. Osteopontin in gingival crevicular fluid. J Periodontal Res. 2001;36:328–33.
Prado-Montes de Oca E, Velarde-Félix JS, Ríos-Tostado JJ, Picos-Cárdenas VJ, Figuera LE. SNP 668C (−44) alters a NF-κB1 putative binding site in non-coding strand of human β-defensin 1 (DEFB1) and is associated with lepromatous leprosy. Infect Genet Evol. 2009;9:617–25.
Ricci E, Malacrida S, Zanchetta M, Montagna M, Giaquinto C, De Rossi A. Role of β-defensin-1 polymorphisms in mother-to-child transmission of HIV-1. J Acquir Immune Defic Syndr. 2009;51:13–9.
Kocsis AK, Kiss ZF, Tiszlavicz L, Tiszlavicz Z, Mándi Y. Potential role of human β-defensin 1 in Helicobacter pylori-induced gastritis. Scand J Gastroenterol. 2009;44:289–95.
Tiszlavicz Z, Szabolcs A, Takács T, Farkas G, Kovács-Nagy R, Szántai E, Sasvári-Székely M, Mándi Y. Polymorphisms of β defensins are associated with the risk of severe acute pancreatitis. Pancreatology. 2010;10:483–90.
Ozturk A, Famili P, Vieira AR. The antimicrobial peptide DEFB1 is associated with caries. J Dent Res. 2010;89:631–6.
Azevedo LF, Pecharki GD, Brancher JA, Cordeiro CA Jr, Medeiros KG, Antunes AA, Arruda ES, Werneck RI, de Azevedo LR, Mazur RF, Moysés SJ, Moysés ST, Faucz FR, Trevilatto PC. Analysis of the association between lactotransferrin (LTF) gene polymorphism and dental caries. J Appl Oral Sci. 2010;18:166–70.
Boniotto M, Hazbón MH, Jordan WJ, Lennon GP, Eskdale J, Alland D, Gallagher G. Novel hairpin-shaped primer assay to study the association of the -44 single-nucleotide polymorphism of the DEFB1 gene with early-onset periodontal disease. Clin Diagn Lab Immunol. 2004;11:766–9.
Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, Tack BF, McCray PB Jr. Production of β-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–5.
Krisanaprakornkit S, Weinberg A, Perez CN, Dale BA. Expression of the peptide antibiotic human β-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect Immun. 1998;66:4222–8.
Kalus AA, Fredericks LP, Hacker BM, Dommisch H, Presland RB, Kimball JR, Dale BA. Association of a genetic polymorphism (−44 C/G SNP) in the human DEFB1 gene with expression and inducibility of multiple β-defensins in gingival keratinocytes. BMC Oral Health. 2009;9:21.
Naslavsky MS, Rocha CR, Lima Filho JL, Crovella S. Predicting alternative candidates as binding sites to DEFB1 668 (−44) SNP: a long way from statistical association with multifactorial diseases. Infect Genet Evol. 2009;9:1129–31.
Naslavsky MS, Crovella S, Lima Filho JL, Rocha CR. The sound of silence: human β-defensin-1 gene untranslated SNPs change the predicted mRNA secondary structure in a length-dependent manner. Immunol Lett. 2010;129:53–5.
Ford PJ, Gamonal J, Seymour GJ. Immunological differences and similarities between chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:111–23.
Dommisch H, Açil Y, Dunsche A, Winter J, Jepsen S. Differential gene expression of human β-defensins (hBD-1, -2, -3) in inflammatory gingival diseases. Oral Microbiol Immunol. 2005;20:186–90.
Bissell J, Joly S, Johnson GK, Organ CC, Dawson D, McCray PB Jr, Guthmiller JM. Expression of β-defensins in gingival health and in periodontal disease. J Oral Pathol Med. 2004;33:278–85.
Vardar-Sengul S, Demirci T, Sen BH, Erkizan V, Kurulgan E, Baylas H. Human β defensin-1 and -2 expression in the gingiva of patients with specific periodontal diseases. J Periodontal Res. 2007;42:429–37.
Jordan WJ, Eskdale J, Lennon GP, Pestoff R, Wu L, Fine DH, Gallagher G. A non-conservative, coding single-nucleotide polymorphism in the N-terminal region of lactoferrin is associated with aggressive periodontitis in an African–American, but not a Caucasian population. Genes Immun. 2005;6:632–5.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research (No. 21592626) from the Japan Society for the Promotion of Science (JSPS). The authors report no conflicts of interest related to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ikuta, T., Inagaki, Y., Tanaka, K. et al. Gene polymorphism of β-defensin-1 is associated with susceptibility to periodontitis in Japanese. Odontology 103, 66–74 (2015). https://doi.org/10.1007/s10266-013-0139-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10266-013-0139-9