Abstract
Ammonium is the primarily nitrogen source for plant growth, but the molecular basis of ammonium acquisition in fruit species remains poorly understood. In this study, we report on the characterization of two new ammonium transporters (AMT) in the perennial tree Pyrus betulaefolia. In silico analyses and yeast complementation assays revealed that both PbAMT1;3 and PbAMT1;5 can be classified in the AMT1 sub-family. The specific expression of PbAMT1;3 in roots and of PbAMT1;5 in leaves indicates that they have diverse functions in ammonium uptake or transport in P. betulaefolia. Their expression was strongly influenced by ammonium availability. In addition, the transcript level of PbAMT1;5 was significantly affected by the diurnal cycle and senescence hormones. They conferred the ability to uptake nitrogen to the yeast strain 31019b; however, the 15NH4 + uptake kinetics of PbAMT1;3 were different from those of PbAMT1;5. Indeed, PbAMT1;3 had a higher affinity for 15NH4 +, and pH changes were associated with this substrates’ transport in yeast. The present study provides basic gene features and transcriptional information for the two new members of the AMT1 sub-family in P. betulaefolia and will aid in decoding the precise roles of AMTs in P. betulaefolia physiology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ammonium is an important mineral nitrogen source for plants. One kind of integral membrane proteins, ammonium transporters (AMTs), work as the primary mediators of ammonium acquisition and metabolism throughout plants (Loqué and von Wirén 2004; Ludewig et al. 2007). AMTs are encoded by two distinct gene families (AMT1 and AMT2) that do not share a significant overall sequence similarity (Loqué and von Wirén 2004). Both AMT1 and AMT2 subfamilies contain 11 putative transmembrane domains (TMs) and one AMT signature motif (Couturier et al. 2007; McDonald et al. 2012). However, AMT1 proteins possess their own unique characteristics. For example, AMT1-type genes lack introns in the genome sequences, with the exception of LjAMT1;1 (Salvemini et al. 2001). Meanwhile, AMT1 homologs are highly selective for ammonium and can transport methylamine (Couturier et al. 2007), which makes studying their functions convenient. Therefore, a series of higher plant AMT1 sub-family members have been characterized in heterogenous expression system since the first AMT1 gene was identified from Arabidopsis thaliana (Couturier et al. 2007; D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Ludewig et al. 2002; Mayer et al. 2006; Ninnemann et al. 1994; Salvemini et al. 2001; Selle et al. 2005; Shelden et al. 2001; Sonoda et al. 2003; Yuan et al. 2007).
Plant AMT1 proteins appear to have a variety of transport mechanisms depending on their physiological roles (Ludewig 2006). Until now, ammonium uptake-defective yeast complementation and electrophysiological measurement in Xenopus oocytes have been two universal techniques for assessing AMT1 functions (Couturier et al. 2007; D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Ludewig et al. 2002; Ortiz-Ramirez et al. 2011; Sogaard et al. 2009). AMT1-type proteins can be divided into high and relatively low affinity subgroups, which have K m values for ammonium of < 15 μM or over 20 μM, even reaching 317 μM, respectively (D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Gu et al. 2013; Ludewig et al. 2002, 2003; Neuhäuser et al. 2007; Shelden et al. 2001; Sogaard et al. 2009; Yuan et al. 2007). More recently, the complex mechanism of proton regulation of NH4 + transport mediated by AMT1 proteins was uncovered. For example, PvAMT1;1 in Phaseolus vulgaris work as a H+/NH4 + symporter strongly depends on the external pH (Ortiz-Ramirez et al. 2011). The activity of TaAMT1;1 from Triticum aestivum is stimulated by acidic pH; however, adequate proof for proton-reliant adjustments is still absent (Sogaard et al. 2009). In contrast, the currents of the two AMT1 genes from tomato are not affected by changes in the external proton concentration (Ludewig et al. 2002, 2003). Thus, different AMT1 proteins from various species may have their own distinctive characteristics, which help plants adapt to diverse environments.
In addition, multiple forms of AMT1 transporters allow for a greater regulatory flexibility and organelle-, cell-, tissue- or organ-specialization, and enable cells to take up ammonium over a wide range of concentrations (Couturier et al. 2007; D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Gu et al. 2013; Ludewig et al. 2007; Straub et al. 2014; Yuan et al. 2007). Indeed, most AMT1 transporters are highly regulated by nitrogen availability, but their expression patterns display multiformity when plants are grown in different environments. So far AMT expression has been amply studied in several plants, including A. thaliana (Gazzarrini et al. 1999; Ninnemann et al. 1994), Lycopersicon esculentum (Becker et al. 2002; Lauter et al. 1996; von Wirén et al. 2000), Oryza sativa (Sonoda et al. 2003), Lotus japonicus (D’Apuzzo et al. 2004) and Populus tremula × alba (Couturier et al. 2007). Most of the AMT1 genes were preferentially expressed in roots (Couturier et al. 2007; Selle et al. 2005; Shelden et al. 2001; Sonoda et al. 2003; von Wirén et al. 2000), whereas OsAMT1;1 from O. sativa and AtAMT1;1 from Arabidopsis were found in various organs (Shelden et al. 2001; Sonoda et al. 2003). Our previous work showed that PbAMT1;1, one of AMT1-type gene from Pyrus betulaefolia, was detectable in all parts of the seedlings and strongly responded to nitrogen regimes, diurnal cycles and phytohormones (Li et al. 2015). Because the AMT1 sub-family is a small multi-gene family, isolating new AMT1 genes and analyzing their expression patterns and functional characterizations could help in presenting a comprehensive foundation for future studies on the ammonium nutrition of P. betulaefolia.
In this study, two new members of AMTs identified from P. betulaefolia were further functionally characterized. Their gene expression patterns were analyzed under different nitrogen availability in young seedlings. Furthermore, their physiological functions and nitrogen uptake abilities were analyzed in yeast through heterogeneous complementation. The experiments suggest specific expression localizations for PbAMT1;3 and PbAMT1;5, which were detected exclusively in roots and leaves, respectively, rather than in the whole seedling like PbAMT1;1. The 15NH4 + uptake kinetics of the PbAMT1;3 high affinity ammonium transporter in the yeast system were different from those of PbAMT1;5. Specifically, PbAMT1;3 had an obviously higher affinity, contrasting transcriptional regulation with nitrogen availability, and pH changes were associated with substrate transport in yeasts. Because little is known about AMT-type transporters in pear, the gene features of three PbAMT1s (PbAMT1;1, PbAMT1;3 and PbAMT1;5) were compared, and the possible roles of each transporter in P. betulaefolia physiology was discussed.
Materials and methods
Plant material and treatments
Mature seeds of P. betulaefolia were surface sterilized, laminated at 4 °C for approximately 40 days, and then sowed into the culture dish for germination. After that, the seedlings were transferred to the hydroponic system at 23 °C, with 70 % relative humidity, 300 μmol m−2 s−1 light intensity and a 16/8 h light/dark period (Li et al. 2015). The nutrient solution was renewed every 3 days, which contained 0.75 mM CaCl2·2H2O, 4.7 mM KCl, 2.5 mM NH4Cl, 0.375 mM MgSO4·7H2O, 0.3 mM KH2PO4, 100 μM H3BO3, 5 μM KI, 100 μM MnSO4·H2O, 30 μM ZnSO4·H2O, 1 μM Na2Mo4·2H2O, 0.1 μM CuSO4·5H2O, 0.1 μM CoCl2, 100 μM FeSO4·7H2O and 100 μM Na2EDTA·2H2O. When these seedlings were 5 months old, they were collected for different treatments. Five treatments were designed as follows: First group: the seedlings were grown in a nutrient solution containing 2.5 mM NH4Cl, and were subsequently used for the analysis of gene expression levels in various tissues. Second group: the seedlings were grown in a nutrient solution containing 10 mM NH4Cl, which was defined as the high nitrogen treatment. Third group: the seedlings were grown in a nutrient solution deprivation NH4 + for 72 h as nitrogen starvation, and then 2.5 mM NH4Cl was replenished as nitrogen resupply. Fourth group: the seedlings were grown in the normal nutrient solution with a 16 (8:00–24:00)/8 h (0:00–8:00) light/dark period and harvested every 4 h for the diurnal rhythm study. Fifth group: 100 µM abscisic acid (ABA) or 100 μM methyl jasmonate (MeJ) was fed through the roots of the seedlings for 24 h as a phytohormone treatment. A mock treatment with 2.5 mM NH4Cl for 24 h was used as the control (Li et al. 2015). Finally, root, stem and leaf samples from the above five treatments were collected at each time point and frozen in liquid nitrogen for further research.
Gene identification and bioinformatics analysis
Firstly, two transcripts, Pbr000917 and Pbr042281, were found in the roots and leaves, respectively, of transcriptome databases from P. betulaefolia nitrogen-treated seedlings. Then, their nucleotide sequences were employed as electronic probes to search the Pyrus × bretschneideri (Chinese white pear) genome sequencing database at the National Center for Biotechnology Information. Two sequences described as different ammonium transporter 1 member 3-like genes (XM_009373231.1 and XM_009375975.1) were obtained. Therefore, our transcripts, Pbr000917 and Pbr042281, containing complete coding regions were named as PbAMT1;3 and PbAMT1;5, respectively.
Translated protein sequences from PbAMT1;3 and PbAMT1;5 were obtained through BioXM 2.6, and then this software was applied to perform multiple alignments. TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM/) and ScanProsite tools (http://expasy.org/tools/scan-prosite/) were used to find TMs or AMT signature motifs, respectively. To build a neighbor-joining phylogenetic tree, 27 AMT amino acid sequences were aligned using ClustalW and imported into the Molecular Evolutionary Genetics Analysis (MEGA) package version 5.01 (Tamura et al. 2011). After that, the Bootstrap tests were conducted using 1,000 replicates. Branch lengths are proportional to phylogenetic distances.
Quantitative real-time PCR (qPCR)
Total RNA was extracted using a StarSpin Plant RNA Mini Kit (GenStar, Beijing, China), and DNase I (GenStar) was used to eliminate DNA contamination from all RNA samples. Then, first-strand cDNA was synthesized using an M-MLV RTase cDNA Synthesis Kit (TaKaRa, Shiga, Japan). Specific primers for PbAMT1;3 (5′-TCG CTT TTA CGC AGA TTA C-3′/5′-TAC AAT ACT TGC AGT AGG T-3′) and PbAMT1;5 (5′-CTC TCT TCT ACA GCC TTC AC-3′/5′-CGA ATT TCA ATC ATA GCT TG-3′) were designed based on the full length cDNA sequences of their corresponding pear AMT1 members (XM_009373231.1 and XM_009375975.1, respectively). Then, qPCR was carried out to detect the expression levels of PbAMT1;3 and PbAMT1;5 under different treatments. The Pbactin gene was chosen as the internal control in this experiment and qPCR was run on a CFX96 system (Bio-Rad, CA, USA) using a previously reported reaction program and conditions (Li et al. 2015). Three independent biological replicates, each with two technical replicates, were analyzed. A no template reaction was used as a negative control. After normalizing to the transcript abundance of the internal control gene, Pbactin, the expression levels of PbAMT1;3 and PbAMT1;5 were presented as means with standard errors.
Yeast expression and 15NH4 + uptake assays
pYES2 (Invitrogen, USA) plasmids containing the PbAMT1;3 complete coding region as well as PbAMT1;5 were electroporated (MicroPulser, Bio-Rad) into the ura− AMT-defective yeast strain 31019b (triple-Δmep) (Li et al. 2015; Marini et al. 1997), which was kindly provided by Professor Nicolaus Von Wirén (Leibniz Institute for Plant Genetics and Crop Plant Research, Gatersleben, Germany). The nitrogen-deficient growth medium (minimal yeast growth medium without amino acids and ammonium sulfate; Difco, BD, NJ, USA) was used to select positive transformants, which was supplemented with 10 mM arginine, 10, 100 or 500 µM NH4Cl as the sole nitrogen source. 15NH4 + uptake assays were conducted using previously described experimental conditions (Li et al. 2015).
Statistical analysis
Data from at least three independent experiments were analyzed by one-way ANOVA using SigmaPlot v11.1 (Jandel Scientific software, San Rafael, CA, USA), and the differences were compared using Duncan’s test with a significance level of P < 0.05.
Results
Isolation and sequence analyses of PbAMT1s
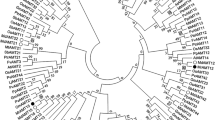
Two cDNAs with complete coding regions were identified from P. betulaefolia through transcriptome sequencing and bioinformatics analyses, which were designated PbAMT1;3 (GenBank accession number KR870986) and PbAMT1;5 (GenBank accession number KR870987). As suggested by a phylogenetic tree based on a protein multiple sequence alignment, three AMT sequences from P. betulaefolia fell in the subgroup AMT1. PbAMT1;3 and PbAMT1;5 were located in the same clade, but PbAMT1;1 belonged to another clade (Fig. 1). An analysis carried out using the TMs prediction program THMM (http://www.cbs.dtu.dk/services/TMHMM/) suggests that the two new AMT1 proteins were likely to have 11 TMs, as do the other AMT plant members (Fig. 2; Couturier et al. 2007; D’Apuzzo et al. 2004; Gazzarrini et al. 1999). An in silico analysis corroborated the results from fusion protein experiments, indicating that the majority of the Mep/Amt proteins possess 11 membrane-spanning helices, with the N-terminus on the exterior face of the membrane and the C-terminus on the interior (Thomas et al. 2000). In addition, Fig. 2 presents a tentative signature sequence for the proteins of PbAMT1;3 and PbAMT1;5, based on the existing AMT signature (McDonald et al. 2012). This sequence [DFAGSGVVHL(M)VGG(A)V(I)AGLWGS(A)F(L)I(V)EGPR] may serve as an identification motif specific to the AMT1 sub-family, and they can be used to identify new AMT1 members in other sequenced plants species (Couturier et al. 2007).
An unrooted phylogenetic tree of the ammonium transporter (AMT) family from plants. Phylogenic analyses were conducted using the neighbor-joining (NJ) method implemented in the Molecular Evolutionary Genetics Analysis (MEGA) package version 5.01. Bootstrap tests were performed using 1,000 replicates. Branch lengths (drawn in the horizontal dimension only) are proportional to phylogenetic distances. The accession numbers of the known AMTs are given as follows: AtAMT1;1 (CAA53473), AtAMT1;2 (AAD54639), AtAMT1;3 (AAD54638), AtAMT1;4 (CAB81458), AtAMT1;5 (NP_189072), AtAMT2 (NP_181363), LeAMT1;1 (X92854), LeAMT1;2 (CAA64475), LeAMT1;3 (Q9FVN0), LjAMT1;1 (AAG24944.1), LjAMT1;2 (AY135020), LjAMT1;3 (AJ575588), LjAMT2;1 (AF187962), PbAMT1;1 (KJ123648), PbAMT1;3 (KR870986), PbAMT1;5 (KR870987), PtrAMT1;1 (XM_002314482), PtrAMT1;2 (XM_002325754), PtrAMT1;3 (XM_002311667), PtrAMT1;4 (XM_002303068), PtrAMT1;5 (XM_002301801), PtrAMT1;6 (XM_002314070), PtrAMT2;1 (XM_002309115), OsAMT1;1 (AF289477), OsAMT1;2 (AF289478), OsAMT1;3 (AF289479), and OsAMT2.1 (AB051864)
Alignment of three s deduced amino acid sequences for PbAMT1s. A blast algorithm was performed with BioXM 2.6 software. Identical residues are shown in black and gaps are indicated as dashes. Transmembrane domains were predicted using the TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM/), and they are outlined by thick lines on the bottom. The AMT signature motif was determined with the ScanProsite tool at ExPaSy (http://expasy.org/tools/scanprosite/) and it is marked by a dotted line with arrows
Expression regulation of PbAMT1;3 and PbAMT1;5
To gain insights into their possible physiological roles, the transcript abundances of the two AMT1-type genes in leaves, stems and roots were investigated. Transcript levels were measured in all of the experiments described previously (in the “Materials and methods” section). The qPCR results indicated that PbAMT1;3 and PbAMT1;5 had distinct transcriptional features in various organs. Indeed, the expression of PbAMT1;3 was restricted to roots; however, the expression of PbAMT1;5 was found mainly in leaves (Fig. 3).
Expression analysis of PbAMT1s in various tissues of P. betulaefolia. The RNA extraction, cDNA acquisition and quantitative real-time PCR operations were carried out as described in the “Materials and methods” section. The transcript levels of PbAMT1s were normalized to Pbactin, which was amplified as an internal control, in the same samples. Values are expressed as means ± standard errors (bars) of three replicate experiments. Different letters indicate significant differences (P < 0.05)
To obtain preliminary information for AMT1 in nitrogen regimes, high nitrogen supply and nitrogen starvation experiments were performed using P. betulaefolia seedlings in a hydroponic system (in “Materials and methods” section). The transcript levels of PbAMT1;3 and PbAMT1;5 in roots and leaves, respectively, were significantly affected after 10 mM NH4Cl was provided or NH4 + was removed from the nutrient solution (Fig. 4). Specifically, a 10 mM NH4Cl treatment induced the transcription of PbAMT1;3 in roots; however, it inhibited the expression of PbAMT1;5 in leaves (Fig. 4a, c). For example, in absorption organs, the abundance of PbAMT1;3 reached 2.45-fold its transcript level in the control roots when the seedlings were exposed to 10 mM NH4Cl at 48 h (Fig. 4a). In contrast, 10 mM NH4 +-supplied plants displayed lower PbAMT1;5 mRNA abundance levels in leaves during the whole treatment process (Fig. 4c). Furthermore, the transcript level of PbAMT1;5 declined to 56.30 % of the non-treated control after the seedlings were grown under high nitrogen conditions for 72 h.
Effect of the nitrogen (N) regime on the expression of PbAMT1s in P. betulaefolia. The RNA extraction, cDNA acquisition and quantitative real-time PCR operations were carried out as described in the “Materials and methods” section. The transcript levels of PbAMT1s were normalized to Pbactin, which was amplified as an internal control, in the same samples. a, b PbAMT1;3 expression in roots under 10 mM NH4Cl supply until 72 h or nitrogen (N) starvation until 72 h, then 2.5 mM NH4Cl was replenished for 8 h. c, d PbAMT1;5 expression in leaves under 10 mM NH4Cl supply until 72 h or nitrogen (N) starvation until 72 h, then 2.5 mM NH4Cl was replenished for 8 h. Values are expressed as means ± standard errors (bars) of three replicate experiments. Different letters indicate significant differences (P < 0.05)
In the case of nitrogen starvation, PbAMT1s transcriptional responses were more complex. Firstly, PbAMT1;3 transcripts strongly increased in roots under the early period of nitrogen starvation (up to 2.37-fold at 48 h), then declined at 72 h, but its transcription was still 1.48-fold of the control (Fig. 4b). After that, 2.5 mM NH4Cl was resupplied and the expression of PbAMT1;3 was slightly inhibited at 2 h, then its transcription mildly increased at 4 h, and finally it returned to 1.23-fold of the control at 8 h (Fig. 4b). Meanwhile, the slight inhibition of PbAMT1;5 transcription was first detected in leaves once nitrogen was eliminated from the solution at 24 h, but this gene revealed the opposite expression tendency in leaves from 48 to 72 h (Fig. 4d). The expression level of PbAMT1;5 declined gradually in leaves when 2.5 mM NH4Cl was replenished to the solution after nitrogen starvation (Fig. 4d).
To investigate the impact of light regimes on the regulation of PbAMT1;5 transcript levels, total RNAs were extracted from leaves of P. calleryana plants grown in a normal solution and collected at 4 h intervals. Figure 5a clearly showed that PbAMT1;5 transcript levels exhibit diurnal rhythms, with the greatest transcript level found at noon (~1.89-fold of the expression of this gene at 4:00 AM). Thus, we may conclude that PbAMT1;5 joins in the transportation of NH4 +/NH3-generated by photorespiration. Additionally, PbAMT1;5 expression was obviously induced after 100 µM ABA or 100 µM MeJ was provided to P. calleryana at 24 h (Fig. 5b). Both ABA and MeJ can trigger leaf senescence; therefore, this AMT1 gene may participate in the nitrogen backflow, especially in the autumn defoliating period.
Effect of the light–dark cycle (a) and phytohormones (b) on PbAMT1;5 expression in leaves of P. betulaefolia. The RNA extraction, cDNA acquisition and quantitative real-time PCR operations were carried out as described in the “Materials and methods” section. The PbAMT1;5 transcript levels were normalized to Pbactin, which was amplified as an internal control, in the same samples. A mock treatment with 2.5 mM NH4Cl for 24 h was used as the control. Values are expressed as means ± standard errors (bars) of three replicate experiments. Different letters indicate significant differences (P < 0.05)
Functional analysis of PbAMT1;3 and PbAMT1;5 in yeast
The complementation experiments with AMT-deficient yeast were effective in identifying the functions of AMT proteins (Couturier et al. 2007; D’Apuzzo et al. 2004; Straub et al. 2014). Here, cells transformed with the empty vector pYES2 or pYES2 harboring PbAMT1;3/PbAMT1;5 were able to grow on the medium containing 1 mM Arg as the sole nitrogen source. Thus, the above three kinds of yeast cells exhibited sufficient activity. Moreover, both transporters were functional, as they complemented the growth on ammonium as the single nitrogen source (Fig. 6). When compared with the empty vector pYES2, both transporters conferred robust yeast growth, which was obviously affected by NH4 + concentration changes between 10 and 500 µM (Fig. 6). Thus, both PbAMT1;3 and PbAMT1;5 encoded functional ammonium transporters.
Functional characterization of PbAMT1;3 and PbAMT1;5 in yeast. Yeast growth on selective media (pH 5.8) with 1 mM arginine (Arg), 10 µM, 100 µM or 500 µM NH4 + as the sole nitrogen source. From top to bottom: control cells with empty plasmid pYES2; cells expressing PbAMT1;3; and cells expressing PbAMT1;5. Pictures were taken after 3 days of growth at 30 °C
Usually, liquid yeast cell cultures have the ability to take up and remove ammonium from the medium; however, the net uptake is saturated at mM levels (Straub et al. 2014). The yeast mutant strain 31019b is defective in the three endogenous NH4 + transporters and can be used as an excellent material for ammonium uptake research (Marini et al. 1997). However, a tiny linear influx of the isotope of 31019b cells carrying pYES2 (the negative control) was also observed during the 15nitrogen-labeled uptake experiments (Fig. 7a–c). This part of the net NH4 + influx may come from the permeation of NH4 + (at pH 4.8–5.8) or the diffusion of NH3 (at pH 6.8) into the yeast cells, which was considered as background uptake thereafter. Commonly, NH4 + and NH3 concentrations were determined from total ammonium concentrations using the Henderson-Hasselbach equation: lg (NH3)/(NH4 +) = pH − pKa, where pKa is 9.25 (Mayer and Ludewig 2006). As expected, the expression of PbAMT1;3 and PbAMT1;5 conferred to yeast 31019b with pYES2 the ability to take up more 15NH4 + in the range of 10–500 µM, in which net 15NH4 + influxes were obtained by subtracting the values of pYES2 under the same 15NH4 + concentration (Fig. 7d–f). Then, kinetic parameters were calculated from a saturable Michaelis–Menten plot. Although PbAMT1;5 also exhibited a high affinity for 15NH4 + (V max ≤ 6.53 pmol NH4 +min−1mg cell at pH 4.8–6.8), its capacity for ammonium transport was at least an order of magnitude lower than that of PbAMT1;3 under the same conditions (Fig. 7d–f; Table 1). Interestingly, the V max of PbAMT1;3 decreased with an increasing external pH, whereas that of PbAMT1;5 was uninfluenced, so that at pH 4.8–6.8 the transport capacity of the two transporters appeared to be dissimilar. The uptake studies indicated that these two AMTs possess disparate affinities for NH4 + and exert opposite responses to external pH changes.
Concentration-dependent ammonium uptake activity of PbAMT1;3 and PbAMT1;5 in yeast. Kinetic analyses were performed as described in the “Materials and methods” section. The net 15NH4 + influxes mediated by PbAMT1;3 and PbAMT1;5 were calculated by subtracting the background values of pYES2. Values are expressed as means ± standard errors (bars) of three replicate experiments. Different letters indicate significant differences (P < 0.05)
Discussion
The AMT1 multigene family in P. betulaefolia
Several points of evidence indicate that proteins of the AMT1 gene family act as functional NH4 + transporters in plants (D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Loqué and von Wirén 2004; von Wirén et al. 2000). After the isolation of PbAMT1;1 (Li et al. 2015), we found that AMT1 is a multigene family consisting of at least three members in P. betulaefolia (Fig. 2). Complete coding region cDNAs were isolated for PbAMT1;3 and PbAMT1;5, and were detected in different tissues (Fig. 3). The existence of several AMT genes in P. betulaefolia emphasizes the importance of NH4 + as a primary mineral nutrient in this species, indicating that NH4 + transport is a tightly regulated process. Indeed, NH4 + is the preferential form for nitrogen uptake at low external concentrations (Gazzarrini et al. 1999; Gu et al. 2013). Therefore, we hypothesized that transporters of the AMT1 gene family are responsible for the large capacity of high-affinity NH4 + uptake in plants. Our results demonstrated that two members of AMT1 (PbAMT1;1 and PbAMT1;3) had a stronger response to NH4 + availability in the roots, whereas, PbAMT1;5 had a stronger response in leaves of P. betulaefolia plantlets (Fig. 4; Li et al. 2015). Furthermore, all of them successfully endowed a yeast AMT mutant 31019b with NH4 + uptake ability (Figs. 6, 7; Li et al. 2015). Thus, we can define their functions in NH4 + uptake and transport.
A comparison of the expression patterns of AMT1 genes in different species might suggest that the transcriptional regulation of this family was complex (Becker et al. 2002; Couturier et al. 2007; Gazzarrini et al. 1999; Lauter et al. 1996; Ninnemann et al. 1994; Selle et al. 2005; Shelden et al. 2001; Sonoda et al. 2003; von Wirén et al. 2000). AMT1 members presumably fulfill discrete, though perhaps partially overlapping, roles for nitrogen absorption and transport in plants (D’Apuzzo et al. 2004). The analysis of PbAMT1 gene expression patterns in different tissues revealed organ specificity and a unique regulation of PbAMT1;1, PbAMT1;3 and PbAMT1;5. Thus, it is not surprising that PbAMT1;1 and PbAMT1;3 are highly expressed in roots, but the transcript of the former was also found in shoot organs (Fig. 3; Li et al. 2015). However, PbAMT1;5 was exclusively detected in leaves (Fig. 3). In addition, the expression levels of all three PbAMT1 genes were significant influenced by nitrogen supply changes (Fig. 4; Li et al. 2015). The transcript features suggested that PbAMT1;1 and PbAMT1;3 worked together in root NH4 + uptake from the growth medium. In addition, PbAMT1;1, along with leaf-specific PbAMT1;5, also may be recruited for taking up NH3 that diffuses from cells during photorespiration in leaves.
Interestingly, AMT1-type proteins are composed of the high affinity subgroup and the relatively low affinity subgroup, based on their K m for ammonium (D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Gu et al. 2013; Ludewig et al. 2002, 2003; Neuhäuser et al. 2007; Shelden et al. 2001; Sogaard et al. 2009; Yuan et al. 2007). Moreover, 15NH4 + uptake in recombination yeast indicated that PbAMT1;1, PbAMT1;3 and PbAMT1;5 had different NH4 + affinities based on their variant K m values (from 6.35 to 26.84 µM) of (Table 1; Li et al. 2015). Hence, PbAMT1;3 was classified in the high affinity subgroup (K m < 10 µM), while both PbAMT1;1 and PbAMT1;5 were classified in the relatively low affinity subgroup (K m > 20 µM). What is more, the regulatory mechanism of pH values on NH4 + transport in transformed yeast mediated by PbAMT1 proteins was irregular. For example, the V max for 15NH4 + uptake of PbAMT1;3, like PvAMT1;1 and TaAMT1;1, was largely dependent on the external pH (Table 1; Ortiz-Ramirez et al. 2011; Sogaard et al. 2009). However, adequate proof for a proton-reliant adjustment needs to be provided. In contrast, some AMT1 genes, such as PbAMT1;1, PbAMT1;5, LeAMT1;1 and LeAMT1;2 are not affected by changes in the external proton concentration (Table 1; Li et al. 2015; Ludewig et al. 2002, 2003). These results indicate that different AMT1 members from P. betulaefolia may have their own distinctive characteristics, which help pear adapt to multifarious nitrogen surroundings.
The root-specific PbAMT1;3 is highly regulated by nitrogen availability and pH change
Evidence has shown that the transcriptional response of AMT genes to nitrogen changes is a widespread phenomenon among plants (Becker et al. 2002; Couturier et al. 2007; D’Apuzzo et al. 2004; Gazzarrini et al. 1999; Gu et al. 2013; Lauter et al. 1996; Li et al. 2015; Loqué and von Wirén 2004; Ninnemann et al. 1994; Shelden et al. 2001; Sonoda et al. 2003; von Wirén et al. 2000). Here, the expression of PbAMT1;3 displayed a similar rule (Fig. 4a, b). Specifically, its transcripts, like those of other AMT1s (AtAMT1;2, AtAMT1;3; PtrAMT1;2 and OsAMT1;2), were specifically and highly expressed in roots (Fig. 3; Couturier et al. 2007; Gazzarrini et al. 1999; Selle et al. 2005; Shelden et al. 2001; Sonoda et al. 2003), suggesting that these AMT1 genes play roles in root NH4 + uptake from the culture environment.
The expression of PbAMT1;3 tended to reflect the level of nitrogen supplied (Fig. 4a, b). The transcript of PbAMT1;3 was induced in roots once the plantlets were subjected to a high nitrogen environment (Fig. 4a). Like the PbAMT1;3 gene, the transcript level of OsAMT1;2 from O. sativa displays root-specific and ammonium-inducible properties (Sonoda et al. 2003). However, removing nitrogen from the media resulted in an increase in PbAMT1;3 expression during the first 48 h, then its transcript level was approximately restored to the control level by 72 h (Fig. 4b). Other root-specific AMT1 members showed diverse reactions to nitrogen nutrition limitations. For example, the expression of AtAMT1;2 in roots was insensitive to changes in nitrogen nutrition (Shelden et al. 2001). In the case of OsAMT1;2, its transcript level decreased at 1 h and almost disappeared after 4 h once O. sativa was transferred to a nitrogen-deficient solution, whereas the expression levels of AtAMT1;3 and PtrAMT1;2 were enhanced under the same conditions (Couturier et al. 2007; Gazzarrini et al. 1999; Sonoda et al. 2003). Resupplying NH4 + to the roots led to a slight decrease in PbAMT1;3 expression levels but the augmentation of PtrAMT1;2 transcripts (Fig. 4b; Couturier et al. 2007). By contrast, the expression of OsAMT1;2 relied heavily on the ammonium dose provided following a nitrogen-free treatment (Sonoda et al. 2003). In general, root-specific AMT1 transporter genes from different species show a distinct regulation in response to nitrogen, which might suggest that plant species that grow in various environments or with different life styles organize AMT with orthologous AMT1 members performing their fairly unique functions (Couturier et al. 2007; McDonald et al. 2012).
The balance between NH4 + and NH3 is usually determined by the pH of an aqueous solution. When the pH is neutral, more than 99 % of ammonia is composed of protonated NH4 + (Britto et al. 2001). Once the pH reaches 7.0, only about 1 % NH4 + is lost and approximately 30-fold of NH3 was detected (Britto et al. 2001). Thus, the investigation of the pH profiles of AMT activity would validly identify whether NH4 + or NH3 is the transported molecule (Britto et al. 2001; Ludewig et al. 2002; Soupene et al. 2001). If NH3 were transported, then the ammonium transport rates unquestionably increase with the increasing pH. However, our data disagreed with this hypothesis. Even PbAMT1;3 activity in yeast exhibited strong responses to changes in the external pH, whereas, the 15NH4 + uptake decreased nearly 21.99 % by a change of pH from 4.8 to 5.8, and reduced by 29.05 % when the pH value increased from 5.8 to 6.8 (Table 1). This phenomenon was similar to the results from wheat TaAMT1;1, bean PvAMT1;1, and Lotus japonicus LjAMT2;2, which were more efficient in their NH4 + uptake under acidic pH environments (Güether et al. 2009; Ortiz-Ramirez et al. 2011; Sogaard et al. 2009). Therefore, we conclude that the transport ability of PbAMT1;3 was dependent on the external pH.
The leaf-specific PbAMT1;5 has distinct responses to various factors
The expression of PbAMT1;5 was exclusively detected in leaf tissues and subject to regulation by nitrogen supplies (Figs. 3, 4). One close ortholog, LeAMT1;3, was also mainly found in leaves (von Wirén et al. 2000), however, PtrAMT1;6, another close ortholog of PbAMT1;5, was expressed in the female flowers and primarily in senescing leaves (Fig. 2; Couturier et al. 2007). Furthermore, the amount of PbAMT1;5 transcript decreased in leaves 24 h after being fed 10 mM NH4Cl. Maintaining this high nitrogen treatment resulted in a decline in PbAMT1;5 transcript levels until 72 h (Fig. 4c). The two orthologs, LeAMT1;3 and PtrAMT1;6, were not influenced by high-concentration NH4 + conditions (Couturier et al. 2007; von Wirén et al. 2000). In the case of nitrogen starvation, the expression level of PbAMT1;5, unlike LeAMT1;3 and PtrAMT1;6 genes, quickly declined at 24 h, and then increased. Additionally, the suppression of this gene’s transcription was observed when 2.5 mM NH4Cl was resupplied following nitrogen deprivation (Fig. 4d). In contrast to the situation in P. betulaefolia, nitrogen starvation down-regulated PtrAMT1;6 mRNA abundance and re-supplying NH4 + had a tiny effect on this gene’s expression in leaves (Couturier et al. 2007). The transcription of LeAMT1;3 was not affected by nitrogen deficiency (von Wirén et al. 2000). These phenomena suggested that the organ-specific expression pattern of the AMT1 sub-family could be regulated by nitrogen requirements and external nitrogen status, which may be involved in the diverse translocational and distributional mechanisms of ammonium ions between herbs (O. sativa or L. esculentum) and woody (P. betulaefolia) plants.
However, PbAMT1;5 transcript levels, like PbAMT1;1, exhibited diurnal rhythms. The greatest diurnal changes in transcript levels were found at noonday when photo-respiration is strongest (Fig. 5a). This suggests that the transcriptional regulation of PbAMT1;1 and PbAMT1;5 might be linked to the diurnal regulation mediated by the circadian clock. In fact, the photo-respiratory nitrogen cycle generates a large amount of NH4 + in leaf mitochondria that is subsequently transported to chloroplasts for reassimilation by glutamine synthetase, implying that the expression of PbAMT1 (PbAMT1;1 and PbAMT1;5) genes can be important to ensure the recycling of NH4 + during photorespiration (D’Apuzzo et al. 2004; Li et al. 2015). At the same time, the transcript levels of two closer orthologs of PbAMT1;5, PtrAMT1;6 and LeAMT1;3, were also strongly affected by the diurnal cycle (Couturier et al. 2007; von Wirén et al. 2000). In fact, PtrAMT1;6 expression correlated well with the sugar content, whereas LeAMT1;3 might be linked to the synthesis of asparagine or to deamination of glutamate. All of the above AMT1 genes displayed variations in diurnal rules, which may be based on their unique physiological functions. In addition, some AMT genes were found to assimilate nitrogen during leaf senescence (Couturier et al. 2007; van der Graaff et al. 2006). Here, PbAMT1;5, like PbAMT1;1, was specifically and highly expressed in leaves once the seedlings encountered aging-related hormones (ABA and MeJ) (Fig. 5b; Li et al. 2015). Our data suggest that AMT1 members from P. betulaefolia might also be recruited to ensure ammonium assimilation in senescing leaves during autumn.
Protons are a major factor that adjusts the NH4 + transport function of AMT1 proteins, and their regulatory mechanism is more complex. Once the external pH changes, accompanying proton concentrations vary, and different AMT1 genes show distinct performances. For instance, the K m values of LeAMT1;1, LeAMT1;2 and PbAMT1;1 were independent of the extracellular pH (Li et al. 2015; Ludewig et al. 2002; Mayer et al. 2006). In this study, the NH4 +-induced current of PbAMT1;5 was unaffected by changes in the external proton concentration (Fig. 7; Table 1), excluding NH3 (Soupene et al. 2001) or H+/NH4 + co-transport (Britto et al. 2001) as a transport mechanism and supporting the uniport of charged NH4 + (Ludewig et al. 2002). However, the NH4 + uptake of PvAMT1;1, TaAMT1;1 and PbAMT1;3 was largely dependent on the external pH (Table 1; Ortiz-Ramirez et al. 2011; Sogaard et al. 2009). This indicted that multiple AMT genes with different regulatory mechanisms allow the plant to respond flexibly to diverse nutritional conditions in the environment (Couturier et al. 2007).
Conclusion
In summary, the data presented here on the specific expression patterns of two new PbAMT1 genes and their biochemical properties indicates that each transporter plays a distinct role in P. betulaefolia. Indeed, our study provides a primary foundation for future genetics experiments that will help to illuminate the definite roles of each transporter.
Abbreviations
- ABA:
-
Abscisic acid
- AMT:
-
Ammonium transporter
- Arg:
-
Arginine
- MEGA:
-
Molecular evolutionary genetics analysis
- MeJ:
-
Methyl jasmonate
- OD600 :
-
Absorbance value at 600 nm
- qPCR:
-
Quantitative real-time PCR
- TMs:
-
Transmembrane domains
References
Becker D, Stanke R, Fendrik I, Frommer WB, Vanderleyden J, Kaiser WM, Hedrich R (2002) Expression of the NH4 +-transporter gene LEAMT1;2 is induced in tomato roots upon association with N2-fixing bacteria. Planta 215:424–429
Britto DT, Glass ADM, Kronzucker HJ, Siddiqi MY (2001) Cytosolic concentrations and transmembrane fluxes of NH4 +/NH3. An evaluation of recent proposals. Plant Physiol 125:523–526
Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol 174:137–150
D’Apuzzo E, Rogato A, Simon-Rosin U, El Alaoui H, Barbulova A, Betti M, Dimou M, Katinakis P, Marquez A, Marini AM, Udvardi MK, Chiurazzi M (2004) Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation spatial expression. Plant Physiol 134:1763–1774
Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11:937–948
Gu R, Duan F, An X, Zhang F, von Wirén N, Yuan L (2013) Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant Cell Physiol 54:1515–1524
Güether M, Neuhaüser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P (2009) A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol 150:73–83
Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB (1996) Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. PANS 93:8139–8144
Li H, Cong Y, Lin J, Chang YH (2015) Molecular cloning and identification of an ammonium transporter gene from pear. Plant Cell, Tissue Organ Cult 120:441–451
Loqué D, von Wirén N (2004) Regulatory levels for the transport of ammonium in plant roots. J Exp Bot 55:1293–1305
Ludewig U (2006) Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus Clin Biol 13:111–116
Ludewig U, von Wirén N, Frommer WB (2002) Uniport of NH4 + by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem 277:13548–13555
Ludewig U, Wilken S, Wu BH, Jost W, Obrdlik P, El Bakkoury M, Marini AM, Andre B, Hamacher T, Boles E, Frommer WB, von Wirén N (2003) Homo- and hetero-oligomerization of ammonium transporter-1 NH4 + uniporters. J Biol Chem 278:45603–45610
Ludewig U, Neuhduser B, Dynowski M (2007) Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett 581:2301–2308
Marini AM, SoussiBoudekou S, Vissers S, Andre B (1997) A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol 17:4282–4293
Mayer M, Ludewig U (2006) Role of AMT1;1 in NH4 + acquisition in Arabidopsis thaliana. Plant Biol 8:522–528
Mayer M, Dynowski M, Ludewig U (2006) Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem J 396:431–437
McDonald TR, Dietrich FS, Lutzoni F (2012) Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: toward a new functional and evolutionary classification. Mol Biol Evol 29:51–60
Neuhäuser B, Dynowski M, Mayer M, Ludewig U (2007) Regulation of NH4 + transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 143:1651–1659
Ninnemann O, Jauniaux JC, Frommer WB (1994) Identification of a high-affinity NH4 + transporter from plants. EMBO J 13:3464–3471
Ortiz-Ramirez C, Mora SI, Trejo J, Pantoja O (2011) PvAMT1;1, a highly selective ammonium transporter that functions as H+/NH4 + symporter. J Biol Chem 286:31113–31122
Salvemini F, Marini AM, Riccio A, Patriarca EJ, Chiurazzi M (2001) Functional characterization of an ammonium transporter gene from Lotus japonicus. Gene 270:237–243
Selle A, Willmann M, Grunze N, Gessler A, Weiss M, Nehls U (2005) The high-affinity poplar ammonium importer PttAMT1.2 and its role in ectomycorrhizal symbiosis. New Phytol 168:697–706
Shelden MC, Dong B, de Bruxelles GL, Trevaskis B, Whelan J, Ryan PR, Howitt SM, Udvardi MK (2001) Arabidopsis ammonium transporters, AtAMT1;1 and AtAMT1;2, have different biochemical properties and functional roles. Plant Soil 231:151–160
Sogaard R, Alsterfjord M, MacAulay N, Zeuthen T (2009) Ammonium ion transport by the AMT/Rh homolog TaAMT1;1 is stimulated by acidic pH. Pflug Arch Eur J Physiol 458:733–743
Sonoda Y, Ikeda A, Saiki S, von Wirén N, Yamaya T, Yamaguchi J (2003) Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol 44:726–734
Soupene E, Ramirez RM, Kustu S (2001) Evidence that fungal MEP proteins mediate diffusion of the uncharged species NH3 across the cytoplasmic membrane. Mol Cell Biol 21(17):5733–5741
Straub D, Ludewig U, Neuhäuser B (2014) A nitrogen-dependent switch in the high affinity ammonium transport in Medicago truncatula. Plant Mol Biol 86:485–494
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thomas GH, Mullins JG, Merrick M (2000) Membrane topology of the Mep/Amt family of ammonium transporters. Mol Microbiol 37:331–344
van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792
von Wirén N, Lauter FR, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB (2000) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J 21:167–175
Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N (2007) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19:2636–2652
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Number: 31372051).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, H., Han, JL., Chang, YH. et al. Gene characterization and transcription analysis of two new ammonium transporters in pear rootstock (Pyrus betulaefolia). J Plant Res 129, 737–748 (2016). https://doi.org/10.1007/s10265-016-0799-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0799-y