Abstract
The patterns of homologue segregation are the basis for euploidy or aneuploidy formation in diploids and allo-/auto-polyploids. Homologue segregation in diploids resembles that in allopolyploids during meiosis; however, meiotic chromosome behavior in autopolyploids is complicated by multiplication of homologous chromosome components. Obviously, loss of single chromosomes (or segmented chromosomes) frequently leads to abortion of reproductive gametes in diploids and allopolyploids. In contrast, the consequence of chromosome loss in autopolyploids is effortlessly compensated for by the presence of multiplied chromosome complements. Here, we use the meiotically asynaptic gene asy1, in combination with polyploidization, to elucidate aneuploidy formation in autotetraploid Arabidopsis. The results indicate that, due to homologous asynapsis in meiotic prophase I, retarded chromosome losses could induce aneuploidy during gametogenesis in autotetraploid asy1. The severe loss of individual chromosomes probably reaches the haploid genome among selfed or backcrossed progeny, leading to stochastic chromosome loss in Arabidopsis. Reciprocal crosses of autotetraploid asy1 with wild-type prove a pathway of duoparental transmission of aneuploidy (hypoploidy and hyperploidy). Viable hypoploids over-transmit via male gametes; conversely, viable hyperploids transmit mainly in female gametogenesis. This result suggests a more stringent maternal restriction of ploidy transmission in autopolyploid Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Change in ploidy is a prominent process in plant speciation and genome diversity. Approximately 70% of flowering plants have undergone polyploidization during evolution (Ramsey and Schemske 1998; Soltis et al. 2008). In plant sexual cycles, the chromosome set is reduced by half meiotically due to the fact that there are two rounds of nuclear division to every one round of DNA replication producing haploid gametes, unequivocally demonstrating that ploidy alternation of chromosome sets (2n → n) dominates plant reproductive life cycles.
Based on nuclear chromosome sets, the ploidy of flowering plants can be categorized into three distinct groups: euploid, hypoploid and hyperploid; hypoploidy and hyperploidy together are termed aneuploidy. Cytogenetically, euploid sets of chromosomes arise from equal segregation of homologous chromosomes under the strict surveillance of meiotic genes. The malfunction of this machinery can lead to fatal improper segregation of homologous chromosomes and produce aneuploid progeny via microspore cells or embryo-sac cells.
The drastic effects of imbalanced gene-dosage on phenotype (e.g., aborted pollen with reduced fertility) in aneuploid plants are apparently more severe than those in polyploid plants (Birchler et al. 2007; Huettel et al. 2008). Phenotypic effects could be interpreted by the plausible hypothesis that the dosage imbalance of genes encoding regulatory molecules and/or transcription factors disturbs the stoichiometry of multi-component regulatory complexes and eventually disrupts normal cellular processes (Birchler et al. 2007). Typically, the dosage imbalance involves the products of house-keeping genes and metabolic processes. In contrast, polyploid plants, with their increased dosage of overall genome, exhibit some specific changes in phenotype but seldom exhibit the extreme effects observed in aneuploid plants. Contrasting with the effects caused by polyploidization, aneuploidization engenders more negative than positive effects (Guo and Birchler 1994). For instance, in comparison with diploid plants such as Arabidopsis, tetraploids have reduced fertility and vigorous development, while aneuploids usually exhibit severe infertility and some developmental defects. The aneuploidy induced by segmental chromosomes in maize is associated with severe phenotypic syndromes including reduced stature, tassel alterations and knot transplacement (Makarevitch et al. 2008), and, where gene expression was examined, it was also found to be affected. Besides, the copy number of genes on individual chromosome can be multiplied by aneuploidization. Specifically, phenotypes are regulated quantitatively, and multiplying a single allele can contribute to a phenotypic alternation. Thus, aneuploidization can facilitate mapping of quantitative trait loci. The effects of aneuploidy on genomic structure and epigenetic changes have been documented (Mittelsten Scheid et al. 1996; Papp et al. 1996; Matzke et al. 2003; Huettel et al. 2008). Aneuploidy-associated structural rearrangement was triggered by chromosomal imbalance, and epigenetic alteration also occurred in specific genotypes.
Although recent investigations have referred to aneuploidy and its genetic variation (Henry et al. 2005), as well as the effects of aneuploidy on genome structure and expression (Huettel et al. 2008), so far there have been few studies on aneuploidy transgeneration and its underlying mechanism in polyploid Arabidopsis in comparison with wealth of information available on the role of polyploidization in plant speciation. Using asynaptic gene asy1 (Caryl et al. 2000), the present study aims to characterize aneuploidy transmission and its cytogenetic role in polyploid Arabidopsis. In practice, we hope our work will provide a useful tool for plant breeding through utilizing or avoiding aneuploidy.
Materials and methods
Plant materials and growth conditions
Diploid wild-type and heterozygous asy1 Arabidopsis thaliana Columbia ecotype (Col) were obtained from the Arabidopsis Biological Resources Center (ABRC). All plants used were grown in a growth chamber provided with compound light systems of white and fluorescent lamps at 22°C with a 16/8 h light/dark alternating photoperiod.
Plant tetraploidization and flow cytometric analysis
Tetraploid A. thaliana (wild-type and heterozygous asy1) were generated as follows: diploid Arabidopsis seeds were surface sterilized with 13%(w/v) bleach solution (sodium hypochlorite), sown on 1/2 MS agar medium (Murashige and Skoog 1962) supplemented with 0.5% sucrose, stratified for 4 days at 4°C in the dark to relieve seed dormancy, then moved to 16/8 h light/dark, and grown for 2 weeks. Seedlings on the plate were submerged for 2 h in 0.1% (w/v) colchicine solution in darkness. The treated seedlings were washed with copious amounts of fresh tap water to remove the residual chemical solution, and then gently transplanted into moist soil. Seeds were collected from individual plants and plumpish candidates were selected under a stereomicroscope and sown on moist soil. Nuclear genome content of these plants was estimated by flow cytometry. Briefly, at the rosette stage, a single leaf was collected and chopped in 250 μl ice-cold nuclei extraction buffer with sharp razor blade in a round Petri dish sitting on a bed of ice until a fine suspension was obtained. After incubating on ice for 30 s to 1 min, 750 μl ice-cold Cy-Stain (Partec) solution was added to stain nuclei with fluorescent dye, and the suspension was filtered through 50 μm nylon mesh and incubated for 1 min, then analyzed in a flow cytometer with UV light (wavelength λ = 420 nm) for ploidy determination. The ploidy level of unknown samples was compared with internal standards determined by chromosome counting of known diploid and tetraploid plants generated under the same growth conditions.
Genomic DNA extraction and genotyping
Fresh young leaves of individual plants were collected and quickly frozen in liquid nitrogen. Genomic DNA was extracted with Nucleon PhytoPure Genomic DNA Extraction Kits (GE Healthcare) according to the manufacturer's instructions. Tetraploid asy1 plants were selected from among a large selfed population of heterozygous asy1; genotyping was achieved by PCR reactions with the insertional T-DNA primer pairs kan +/kan− and wild-type ASY1-geno-F/ASY1-geno-R: kan + (5′-TTTTGTCAAGACCGACCTGTCC-3′), kan− (5′-ATGCTCTTCGTCCAGATCATCC-3′), ASY1-geno-F (5′-CTCCATTTCGTATTAGCTGT CG-3) and ASY1-geno-R (5′-CTAGTCTACAAGTCGAAATGAGTC-3). PCR reactions were performed using standard procedures: denaturing at 94°C for 30 s; annealing at 55°C for 30 s; elongation at 72°C for 30 s to 1 min; 35 cycles in total.
Cytological analysis
Chromosomes were prepared as described previously by Ross et al. (1997) and Armstrong et al. (2002) with minor modifications. Briefly, whole inflorescences with flower buds of appropriate sizes of 0.5–2.0 mm in length were collected, and fixed in freshly prepared ethanol/acetic acid (3:1) overnight at room temperature. Fixed inflorescences were then rinsed in distilled water (3 × 5 min) and citrate buffer (10 mM sodium citrate, pH 4.5; 3 × 5 min). Subsequently, the flower buds were digested with mixed enzymes in 0.3% (w/v) pectolyase (Sigma), cellulase (Sigma) and cytohelicase (Sepracor) in citrate buffer at 37°C for 3 h. After digestion, the flower buds were transferred to citrate buffer and stored at 4°C until use. Individual flower buds were transferred onto clean slides under a dissecting microscope and chopped to form a homogeneous mixture with addition of a drop (5–7 μl) of 60% acetic acid until transparent. Slides were placed on a hot plate (55°C) for 10 s, a clean glass cover was applied and gently squashed, and the slides were then quickly frozen and the cover removed with a blade. The prepared slides were air-dried and finally stained with 5 μg/ml DAPI (4′, 6′-diamidino-2-phenylindole) solution.
Germination test and chromosome count
To test their viability, aneuploid seeds collected from selfed and crossed tetraploid plants were surface sterilized with 13% (w/v) bleach solution, and sown on solid 1/2 MS agar medium. Meanwhile, root-tips of 3-day-old seedlings were pretreated for 1 h in ethanol/acetic acid (3:1) fixative at room temperature and then rinsed with citrate buffer solution (10 mM, pH 4.5; 3 × 5 min), and then incubated in an enzyme mixture including 0.3% (w/v) pectolyase (Sigma), 0.3% (w/v) cytohelicase (Sepracor), and 0.3% (w/v) cellulase (Sigma) in citrate buffer for 30 min at 37°C. After enzymatic treatment, the root-tips were transferred to 45% acetic acid for 5–10 min until the root-tips turned transparent. Chromosome squash was prepared as previously described by Armstrong et al. (2002) and stained with 10 μg/ml DAPI solution.
Results
Colchicine-induced tetraploidization
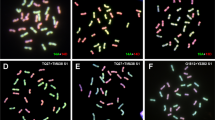
Different methodologies can be used to generate autotetraploid Arabidopsis from diploid progenitors. Polyploidization occurs during plant callus culture in vitro. Although the ploidy levels range between 2 and 15 × (10–75 chromosomes) in Arabidopsis primary tissue culture, the frequency of polyploidy cells is nevertheless still affected by culture time (Fras and Maluszynska 2004). Thus direct polyploidization by colchicine treatment was preferred as a simpler and faster procedure. In addition, chromosome behavior was checked after colchicine-induced autotetraploidization in Arabidopsis wild-type background (Weiss and Maluszynska 2000; Santos et al. 2003), which provides an additional reference for analyzing autotetraploid asy1. In the procedure, diploid heterozygous asy1 were submerged in a 1% colchicine solution for 2 h in darkness, and then transferred to soil. The plumpish selfed seeds collected were sown and, at the rosette stage, the genomic contents of individual plants were determined by flow cytometric analysis (Fig. 1a–d). Counting of the chromosomes of the young flower buds at the mitotic prophase confirmed the results of flow cytometric analysis (Fig. 1e). The genotypes of tetraploid candidates were determined by PCR with primer pairs corresponding to wild-type and the T-DNA insertion referred to above.
Flow cytometric analysis for tetraploidy after colchicine treatment (a–d) and chromosome counts (e). In the cytometric analysis, the first gain peak of genomic contents at the tetraploid-level shifts two-fold behind that of the diploid level. a, b Controls in diploid and tetraploid 2 × Col and 4 × Col; c, d the assayed diploid and heterozygous asy1 tetraploid. e Chromosome counting of tetraploid asy1 at prophase of mitosis (20 chromosomes, each chromosome contains two sister chromatids)
Tetraploid asy1 and its fertility
To analyze aneuploidy propagated by tetraploid asy1, the segregating population was chosen from the selfed tetraploid heterozygous asy1 on the basis of Mendel’s law of segregation. Of the tetraploid candidates, 3 out of 113 were identified as tetraploid asy1, and 4 out of 15 diploid candidates as diploid asy1, which fitted well with expected segregation ratios (P value = 0.01, 1:36 and 1:4, respectively). In most phenotypic features, tetraploids resemble diploids in all aspects of their vegetative growth and development in Arabidopsis (Fig. 2); however, owing to doubled genome content, tetraploids have an elongated growth period and increased organ size. Besides, tetraploid seeds appear relatively plumpish and larger than diploids. Conversely, tetraploids have reduced fertility compared to diploids, indicating the likely disrupted viability of pollen and/or embryo sac after tetraploidization. Likewise, diploid and tetraploid asy1 show considerably reduced fertility and much shortened siliques compared with wild-type (Fig. 3). Nevertheless, the tetraploid asy1 exhibits fertility levels similar to that of diploid asy1 in the mutant, in contrast to the wild-type background (Fig. 4). This intriguing observation leads us to hypothesise that polyploids potentially tolerate more mutation than diploids via aneuploidy formation with respect to fertility.
Male meiotic observation in tetraploid asy1
Cytological analysis of diploid asy1 has been described during development of pollen mother cells (Ross et al. 1997) and embryo-sac cells (Armstrong et al. 2002). However, less information is available about meiotic events of this mutation after direct tetraploidization. Furthermore, although descriptions of meiosis in tetraploid Arabidopsis are supported by data from fluorescence in situ hybridization (Weiss and Maluszynska 2000; Santos et al. 2003), the double-folded chromosome complements, as well as the tiny configuration of individual chromosomes render meiotic analysis of autotetraploids intractable and elusive. Nevertheless, in view of the importance of autopolyploidization, the understanding of meiosis in autotetraploids seems absolutely necessary for polyploidy research. Thus, the typical meiotic stages in tetraploid asy1 are elaborated in parallel with those of the wild-type.
In autotetraploid wild-type Arabidopsis, at the onset of meiosis, the indistinguishable homologous chromosomes (chromatins) gradually condense and initiate synapsis from zygotene to pachytene during prophase I (Fig. 5a); at late prophase I, homologous chromosomes align and become distinguishable with bright DAPI-stained (peri-) centromeres after completing condensation (Fig. 5b); at metaphase I, homologous chromosomes behave as quadrivalents (Fig. 5c) and/or bivalents (Fig. 5d) with several entangled chiasmata; at the transition from metaphase I to anaphase I, homologous chromosomes separate equally in opposite directions (Fig. 5e); from metaphase II to anaphase II, coherent sister chromatids separate and are pulled apart gradually by the centromere-associated spindles (Fig. 5f). In contrast, in tetraploid asy1, homologous chromosomes could partially synapse at early prophase I (Fig. 5g); in succession, the respective chromosomes (with two associated sister-chromatids) appear identifiable, but homologous chromosomes fail to align together (Fig. 5h); afterwards, the majority of chromosomes form univalents (Fig. 5i) or exhibit residual bivalents at metaphase I (Fig. 5j); such univalents are often then retained behind as lagged chromosomes at the first cell division (Fig. 5k). Subsequently, in meiosis II, the lagged chromosomes cannot simutaneously separate into four defined nuclei, leading to indefinite loss of chromosomes (Fig. 5l). Normal chromosome behavior was observed in tetraploid wild-type, whereas in tetraploid asy1, chromosome behavior was distorted despite the partial synapsis and bivalent formation, which is probably attributable to the second yeast HOP1 homologue, ASY2 (Caryl et al. 2000). Therefore, male meiosis in tetraploid asy1 differs from that of tetraploid wild-type in aspects of meiotic chromosome (chromatin) behavior such as chromatin synapsis, and alignment and segregation, which thus probably produces the odd numbers of chromosome complements in the male reproductive gametes.
Progressive meiotic stages of autotetraploid Arabidopsis. Tetraploid wild-type (a–f) and asy1 mutant (g–l). In wild type at early prophase I (a), the linear chromatin is completely synapsed (arrow bright-stained linear synapsed chromatin); b condensed chromosomes align in late prophase I (arrow paired bivalents); c–d paired homologous chromosomes show quadrivalents or bivalents (arrows); e at anaphase I, homologues separate equally to opposite poles; f at telophase II, four nuclei with equal numbers of chromosomes eventually form. In contrast, in tetraploid asy1 (g) linear chromatin appears with partial synapsis at prophase I (arrow bright-stained linear synapsed part); h homologous chromosomes fail to align and exhibit as univalents in late prophase I (arrow single asynaptic univalent); i–j at metaphase I, the homologous chromosomes show mainly univalents or residual bivalents (arrows); k at anaphase I, the homologous chromosomes separate into two poles with retarded chromosomes (arrows three lagged chromosomes); l sister chromosomes separate with retarded chromosomes at the transition from metaphase II to anaphase II (arrows four lagged chromosomes)
Female meiosis in different plant species has been observed in recent years. Havekes et al. (1997) found that, in wild-type tomato, the number of chiasmate chromosome arms in female meiosis was slightly higher than in male meiosis. In the completely asynaptic mutant as6, chromosome pairing and chiasma formation were virtually absent in both sexes, but in the partially asynaptic mutant asb, a higher number of chiasmate chromosome arms in female meiosis than in male meiosis was observed. Although similar chromosome behaviors were obtained in male and female meiosis in wild-type Arabidopsis (Ross et al. 1996; Armstrong and Jones 2001), in the asynaptic mutant asy1, the complete failure of bivalent formation reflected the more severe defects in female meiosis than male meiosis (Sanchez Moran et al. 2001; Armstrong and Jones 2001). This result might indicate a product of gametes with different chromosome numbers after male and female meioses in Arabidopsis asy1 mutants.
Analysis of aneuploidy transmission in tetraploid asy1
The diploid asy1 exhibits disrupted synapsis and failure of homologous pairing, engendering mostly univalent formation, causing severely improper segregation of homologous chromosomes during meiosis (Caryl et al. 2000; Armstrong et al. 2002). Fertility observation of diploid asy1 shows a reduced seed set. The seeds of selfed tetraploid asy1 display varying sizes, and appear shrunken or plumpish (Fig. 6b); approximately 55.1% of seeds are shrunken, and roughly 58% of examined seeds fail to germinate. In contrast, the seeds of tetraploid wild-type are mostly stuffed and exhibit a uniform size (Fig. 6a), as well as a higher germination ratio (approximately 90%). Similarly, backcrossed tetraploid asy1 × a wild-type pollinator resembles the status of selfed tetraploid asy1 with regards to seed development and germination. However, an unexpected outcome was that seed development and viability were significantly improved by crossing tetraploid asy1 (as pollen donor) with wild-type (Table 1). Other independent interploidy crosses of diploids and tetraploids show that the viability of triploid seeds depends mostly on the ploidy level of maternal plant (data not shown). That is, a triploid embryo could be developed in both cross directions, whereas the aborted endosperm was frequently examined when the interploidy cross was conducted with a diploid as the maternal plant. This result confirmed the differential parental effects on seed development (Scott et al. 1998).
The viable seeds of the selfed tetraploid asy1 were dissected by chromosome counting of root-tips. The data (Table 2) show that about 80% of seeds analysed have reduced chromosome numbers (< 20), and at least 25% of analyzed progeny show that the chromosome loss can probably amount to a set of haploid genome; about 7% of observed seeds have increased chromosome complements (> 20). In comparison, the data for the tetraploid wild-type show that nearly 90% of seeds (31:35) possess a normal set of chromosomes in spite of the occasional occurrence of chromosome gain or loss (1–2:35), which can probably be attributed to the unique formation of multivalents such as trivalents plus univalents (Santos et al. 2003) leading to the missegregation of the paired chromosomes of such configurations. These results indicate that asynapsis-induced loss of chromosomes determines aneuploidy formation in tetraploid asy1. Moreover, the severe loss of individual chromosomes hints at a possible stochastic event during meiosis.
To identify the source of aneuploidy (hyperploidy and hypoploidy) during gametogenesis, we crossed tetraploid asy1 with wild-type reciprocally. Chromosome counting indicated that about 45% of the backcrossed seeds analysed have an increased or reduced number of chromosomes when tetraploid asy1 was the pollen recipients, but only 15% of observed seeds show a gain of chromosomes. In comparison, about 70% of seeds examined display loss of chromosomes when tetraploid asy1 was chosen as the pollen donor, but approximately 12% of backcrossed seeds show the euploid chromosome number provided that tetraploid asy1 was selected as pollen donors or recipients. As shown in Table 2, these results prove that hyperploids transmit mainly via female gametogenesis, and hypoploids have a duoparental transmission during the reproductive life cycle but a paternal pathway is preferred in Arabidopsis.
Discussion
The development of Arabidopsis as a plant model for genetic studies is attributed to its small size, short life cycle, its selfing mode of propagation, and the availability of its genomic sequence (Goodman et al. 1995). However, regarding a model for analysis of polyploidy in flowering plants, little is known about ploidy transmission on the basis of autopolyploid Arabidopsis. In this study, we sought to characterize the pathway of aneuploidy transmission in polyploid Arabidopsis.
Differential gametic transmission affects inheritance of rust resistance in allohexaploid wheat (Luig 1960). And the transmission of aneuploid gametes is influenced by the competitiveness of pollen tube growth and fertilization after microsporogenesis and megasporogenesis (Boyd et al. 1970). The induced tetraploid of Lolium perenne shows decreasing frequency of aneuploids with higher numbers of chromosomes in advanced generations, and hyperploids are recovered mainly through female transmission (Ahloowalia 1971). However, the transmission of n + 1 gametes in cabbage trisomics proved that the transmission of extra chromosomes in different trisomics varied in both male and female gametes, and it has been speculated that there is an equal chance of extra-chromosome transmission during gametogenesis (Zhang et al. 2008). Analysis of aneuploid progenies from the asynaptic amphidiploid Scilla scilloides (AABB) clarified that there was no preferential chromosome transmission between genome A and B, and that viable pollen grains might determine aneuploid production (Uchino and Tanaka 1995). Our results indicate that the transmission of aneuploids in autotetraploid Arabidopsis occurs in both male and female gametes, with increased hyperploid transmission arising mainly in female (embryo-sac) cells. Of the two autotetraploid Festuca pratensis, Lolium multiflorum produces more hyperploids, but Lolium perenne shows equal yields in hypoploids and hyperploids (Klinga 1986). Euploids were superior to aneuploids and hyperploids were superior to hypoploids in most developmental features' furthermore, aneuploidy had more pronounced effects on reproductive development (Klinga 1986). In contrast, aneuploids in tetraploid ryegrass were morphologically indistinguishable from eu-tetraploids (Ahloowalia 1971). Our results show that tetraploid asy1 aneuploids resemble eu-tetraploids with respect to several morphologies including reduced fertility.
In animals, including humans, aneuploidy can cause miscarriages and mental retardation (Parry et al. 2002; Duesberg 2007). In four different mice trisomic lines, proliferation and metabolic properties were altered by the presence of an additional chromosomes, as well as immortalization (Williams et al. 2008). Likewise, the development of endosperm was arrested by interploidy hybridization, suggesting a potential determinant of disproportionate contributions of paternal/maternal chromosome complements in Arabidopsis (Scott et al. 1998). Alternatively, imprinted genes such as MEDEA have been proven to influence endosperm development (Vinkenoog et al. 2003). In addition, the locus SENSITIVE TO DOSAGE IMBALANCE (SDI) could tolerate more aneuploidy by buffering the effects of dosage imbalance in triploid-derived recombinant inbred lines (Henry et al. 2007). Dilkes et al. (2008) clarified that the viability of interploidy-crossed seeds was regulated by maternally expressed WRKY transcriptional factor TTG2 by sporophytically affecting endosperm cellularization. In the study, we prove that viable aneuploid gametes with varying chromosome content affect the development of seeds after fertilization, and that the disproportionate ratio of parental chromosome complements may contribute to aberration of seed development.
References
Ahloowalia BS (1971) Frequency, origin, and survival of aneuploids in tetraploid ryegrass. Genetica 42:129–138
Armstrong SJ, Jones GH (2001) Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex Plant Reprod 13:177–183
Armstrong SJ, Caryl AP, Jones GH, Franklin FC (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115:3645–3655
Birchler JA, Yao H, Chudalayandi S (2007) Biological consequences of dosage dependent gene regulatory systems. Biochim Biophys Acta 1769:422–428
Boyd WJR, Sisodia NS, Larter EN (1970) A comparative study of the cytological and reproductive behaviour of wheat and triticale subjected to two temperature regimes. Euphytica 19:490–497
Caryl AP, Armstrong SJ, Jones GH, Franklin FC (2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109:62–71
Dilkes BP, Spielman M, Weizbauer R, Watson B, Burkart-Waco D, Scott RJ, Comai L (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6:2707–2720
Duesberg P (2007) Chromosomal chaos and cancer. Sci Am 296:53–59
Fras A, Maluszynska J (2004) The correlation between the chromosome variation in callus and genotype of explants of Arabidopsis thaliana. Genetica 121:145–154
Goodman HM, Ecker JR, Dean C (1995) The genome of Arabidopsis thaliana. Proc Natl Acad Sci USA 24:10831–10835
Guo M, Birchler JA (1994) Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266:1999–2002
Havekes FW, Jong JH, Heyting C (1997) Comparative analysis of female and male meiosis in three meiotic mutants of tomato. Genome 40:879–886
Henry IM, Dilkes BP, Young K, Watson B, Wu H, Comai L (2005) Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170:1979–1988
Henry IM, Dilkes BP, Comai L (2007) Genetic basis for dosage sensitivity in Arabidopsis thaliana. PLoS Genet 3:e70
Huettel B, Kreil DP, Matzke M, Matzke AJ (2008) Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet 4(10):e1000226
Klinga K (1986) Aneuploidy in induced autotetraploid populations of Festuca pratensis, Lolium multiflorum and Lolium perenne. Hereditas 104:121–130
Luig NH (1960) Differential transmission of gametes in wheat. Nature 185:636–637
Makarevitch I, Phillips RL, Springer NM (2008) Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics 9:7
Matzke MA, Mette MF, Kanno T, Matzke AJ (2003) Does the intrinsic instability of aneuploid genomes have a causal role in cancer. Trends Genet 19:253–256
Mittelsten Scheid O, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J (1996) A change of ploidy can modify epigenetic silencing. Proc Natl Acad Sci USA 93:7114–7119
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Papp I, Iglesias VA, Moscone EA, Michalowski S, Spiker S, Park YD, Matzke MA, Matzke AJ (1996) Structural instability of a transgene locus in tobacco is associated with aneuploidy. Plant J 10:469–478
Parry JM, Al-Obaidly A, Al-Walhaib M, Kayani M, Nabeel T, Strefford J, Parry EM (2002) Spontaneous and induced aneuploidy, considerations which may influence chromosome malsegregation. Mutat Res 504:119–129
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501
Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4:551–559
Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FC, Jones GH (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA transformed lines. Chromosome Res 5:551–559
Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH (2001) Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res 9:121–128
Santos JL, Alfaro D, Sanchez-Moran E, Armstrong SJ, Franklin FC, Jones GH (2003) Partial diploidization of meiosis in autotetraploid Arabidopsis thaliana. Genetics 2165:1533–1540
Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125:3329–3341
Soltis DE, Bell CD, Kim S, Soltis PS (2008) Origin and early evolution of angiosperms. Ann N Y Acad Sci 1133:3–25
Uchino A, Tanaka K (1995) Occurrence of aneuploid progenies from an asynaptic amphidiploid of Scilla scilloides (lindley) druce II mechanism of production of the various aneuploid progenies. J Plant Res 108:185–194
Vinkenoog R, Bushell C, Spielman M, Adams S, Dickinson HG, Scott RJ (2003) Genomic imprinting and endosperm development in flowering plants. Mol Biotechnol 25:149–184
Weiss H, Maluszynska J (2000) Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana. Hereditas 133:255–261
Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322:703–709
Zhang CH, Li XF, Shen SX, Yuan H, Xuan SX (2008) Determination of n + 1 gamete transmission rate of trisomics and location of gene controlling 2n gamete formation in Chinese cabbage (Brassica rapa). J Integr Plant Biol 51:29–34
Acknowledgments
We thank the Gregor Mendel Institute of Plant Molecular Biology in Vienna and the Austrian exchange service (ÖAD) for financial support for our research work. We would like to express our gratitude to two anonymous reviewers who provided constructive comments on our manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s10265-009-0304-y
Rights and permissions
About this article
Cite this article
Wei, F., Zhang, GS. Meiotically asynapsis-induced aneuploidy in autopolyploid Arabidopsis thaliana . J Plant Res 123, 87–95 (2010). https://doi.org/10.1007/s10265-009-0262-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-009-0262-4