Abstract
The taxonomy of Bambusoideae is in a state of flux and phylogenetic studies are required to help resolve systematic issues. Over 60 taxa, representing all subtribes of Bambuseae and related non-bambusoid grasses were sampled. A combined analysis of five plastid DNA regions, trnL intron, trnL-F intergenic spacer, atpB-rbcL intergenic spacer, rps16 intron, and matK, was used to study the phylogenetic relationships among the bamboos in general and the woody bamboos in particular. Within the BEP clade (Bambusoideae s.s., Ehrhartoideae, Pooideae), Pooideae were resolved as sister to Bambusoideae s.s. Tribe Bambuseae, the woody bamboos, as currently recognized were not monophyletic because Olyreae, the herbaceous bamboos, were sister to tropical Bambuseae. Temperate Bambuseae were sister to the group consisting of tropical Bambuseae and Olyreae. Thus, the temperate Bambuseae would be better treated as their own tribe Arundinarieae than as a subgroup of Bambuseae. Within the tropical Bambuseae, neotropical Bambuseae were sister to the palaeotropical and Austral Bambuseae. In addition, Melocanninae were found to be sister to the remaining palaeotropical and Austral Bambuseae. We discuss phylogenetic and morphological patterns of diversification and interpret them in a biogeographic context.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are approximately 80–90 genera and about 1,000–1,500 bamboo species in the world (Clark 1995; Dransfield and Widjaja 1995; Judziewicz et al. 1999; Ohrnberger 1999; McClure 1966; Soderstrom and Ellis 1987). Their natural range includes all continents except Antarctica and Europe, from lowlands up to about 4,000 m in altitude. Biogeographically, bamboos can be categorized into two groups, tropical or temperate (Ohrnberger 1999). In woody bamboos, however, molecular evidence (Clark et al. 1995; Ní Chonghaile 2002) has suggested that they could be divided into at least three major lineages. These can be defined as temperate, palaeotropical, and neotropical.
Bamboo classification has been revolutionized by DNA sequence evidence. The first molecular DNA phylogenetic study to include a broad sample of bamboo diversity (Clark et al. 1995) clearly demonstrated the non-monophyly of Bambusoideae s.l. sensu Clayton and Renvoize (1986). Since this landmark paper, several other studies have supported these results including GPWG (2001) and Bouchenak-Khelladi et al. (2008). In these studies several taxa previously classified in Bambusoideae s.l. were resolved as the most outlying lineages within the grass family (these taxa are now recognized at subfamily rank and include Anomochlooideae, Pharoideae, and Puelioideae). Other taxa included in Bambusoideae s.l. sensu Clayton and Renvoize (1986) have also been shown to be more closely related to Pooideae including the tribes Brachyelytreae, Diarrheneae, and Phaenospermatideae (Clark et al. 1995; GPWG 2001; Bouchenak-Khelladi et al. 2008).
In contrast, the Bambusoideae s.s. sensu Clark et al. (1995) and the GPWG (2001) are well supported as monophyletic (Bouchenak-Khelladi et al. 2008). About 90% of Bambusoideae s.s. are woody bamboos (Ohrnberger 1999). Traditional methods have established classifications within the bamboos and allowed detailed floristic work, but these studies are sometimes limited, because of the lack of morphological characteristics (Stapleton 1997). Phylogenetic relationships at tribal and subtribal levels within Bambusoideae are still unclear and several classifications of woody bamboos are incongruent (Clayton and Renvoize 1986; Soderstrom and Ellis 1987; Dransfield and Widjaja 1995; Li 1998; Ohrnberger 1999; Table 1). There is, therefore, a need for taxonomic stability and it is hoped that molecular DNA data may help identify major groupings. Some studies on taxa of woody bamboos based on molecular phylogenetic analyses have been undertaken such as for the one-flowered, determinate, genera of Bambuseae using rpl16 intron sequence data and morphological characteristics (Clark et al. 2007), 16 Asian genera using restriction site mutations of cpDNA (Watanabe et al. 1994), Bambusa using ITS nuclear rDNA sequences (Sun et al. 2005), Chusquea using chloroplast rpl16 sequence data (Kelchner and Clark 1997), Phyllostachys using different techniques such as RFLP markers (Friar and Kochert 1994), RAPD markers (Gielis et al. 1997), ITS nuclear rDNA sequence data and AFLP markers (Hodkinson et al. 2000), and Schizostachyum using GBSSI and trnL-F sequences (Yang et al. 2007). Combined data-set analyses to study phylogenetic relationships among woody bamboos are rare. There is particularly a need for large multi-gene/sequence phylogenetic trees of Bambusoideae. Plastid DNA variation within the subfamily is low in comparison with several of the other grass subfamilies (Ní Chonghaile 2002) and single-gene analyses of plastid DNA have been found to be insufficient to adequately resolve phylogenetic pattern needed for detailed classification. Few phylogenetic studies have attempted to combine sequences. Ní Chonghaile (2002) applied trnL intron, trnL-trnF intergenic spacer, rpl16 intron sequence data, and ITS nuclear rDNA sequences to study relationships among woody bamboos. However her study focussed mainly on temperate woody bamboos.
In this study, representatives from all subtribes of Bambuseae according to Clayton and Renvoize (1986) and Ohrnberger (1999) were sequenced for five plastid DNA regions (trnL intron, trnL-F intergenic spacer (the names of these two regions are hereafter combined and called trnL-F as they are continuous tandemly arranged sections of DNA sequence), atpB-rbcL intergenic spacer, rps16 intron, and matK gene region) for combined analysis (Table 2). The trnL-F and atpB-rbcL regions are commonly used for phylogenetic study of plants (from species to family levels; Soltis and Soltis 1998), while matK is commonly used for species to order levels. Several studies have shown these genes to be useful for phylogenetic study of grasses and bamboos: trnL-F (Hodkinson et al. 2002; Ní Chonghaile 2002; Yang et al. 2007); matK (e.g. Liang and Hilu 1996; Hilu et al. 1999). The atpB-rbcL and rps16 regions have not previously been used to study bamboo phylogenetics. However, rps16 has proven useful for plant molecular systematics both for dicots, for example Caryophyllaceae (Oxelmann et al. 1997), and for monocots, for example Palmae (Asmussen et al. 2000) and Marantaceae (Andersson and Chase 2001). Combined analysis of plastid DNA regions are often useful for improving phylogenetic resolution and support (Reeves et al. 2001; Hodkinson et al. 2007a). Plastid DNA is generally non-recombining and maternally inherited in most angiosperms. Different sequences found on the plastid genome should therefore share the same evolutionary history and provide congruent phylogenetic trees. The justification to combine datasets in the analyses in this study was based on an examination of groupings (and support for these) found in the single-gene analyses (data not shown). No major and well supported incongruences were found between the results from single gene region analyses and it was deemed appropriate to combine datasets (a total evidence approach).
The objective of this study was to resolve major phylogenetic groupings within Bambusoideae s.s. and evaluate the currently used classifications. More specifically, the objective was:
-
1
to define major bamboo groups and assess the monophyly of existing taxa;
-
2
to study molecular variation in different plastid gene sequence regions to assess their usefulness in bamboo phylogenetics;
-
3
to study the relationships of woody bamboos in comparison with other closely related bamboos and grasses and, especially, to examine the relationship between Bambuseae and Olyreae; and
-
4
to assess the monophyly, inter-relationships, and biogeography of taxa within Bambuseae.
Materials and methods
Plant materials
Sixty-four species from three subfamilies sensu Clayton and Renvoize (1986), Bambusoideae, Pooideae, and Panicoideae, were sampled (Table 2). Panicoid grasses were selected as a suitable outgroup because they lie outside the BEP clade (GPWG 2001) and because we were primarily concerned with establishing relationships of taxa within Bambusoideae s.s.. Bambusoideae s.s. are a robust clade with high levels of support in most recent analyses (Hilu et al. 1999; Zhang 2000; GPWG 2001; Salamin et al. 2002; Bouchenak-Khelladi et al. 2008). We also wanted to determine the closest relatives to Bambusoideae s.s. from within the BEP clade. For this reason an outgroup that outlies the BEP clade was required. Because of the incongruence between infra-subfamilial classifications of Bambusoideae s.s., representatives from all bamboo subtribes according to Clayton and Renvoize (1986) and Ohrnberger (1999) were included. The number of accepted species included per genus (Clayton et al. 2008) is shown in Table 2. For genera not recognized in the website, species number follows Ohrnberger (1999). Phuphanochloa is a bamboo genus new to science (S. Sungkaew et al., in press). Plant material was collected in silica gel to rapidly desiccate the material and minimize DNA degradation (following Chase and Hills 1991). Some samples, however, were obtained from herbarium specimens.
Isolation of total genomic DNA
Total genomic DNA (tDNA) was extracted from ca 0.2 g of leaf using the modified 2XCTAB method of Doyle and Doyle (1987) as outlined in Hodkinson et al. (2007b) and precipitated in isopropanol for at least 1 week or longer at −20°C (Hodkinson et al. 2007b). The tDNA was then pelleted, washed with 70% ethanol, and purified using a JetQuick PCR product-purification kit (Genomed). All DNA samples were processed in this study, except for that from Oreobambos buchwaldii, which was processed by Ní Chonghaile (2002). DNA was then stored in TE buffer (10 mM Tris-HCl; 1 mM EDTA; pH 8.0) at −20°C until use.
DNA amplification and sequencing
The polymerase chain reaction (PCR) was used to amplify each of the five gene regions. Using the primers “c” and “f” designed by Taberlet et al. (1991), the PCR amplification protocol of trnL-F consisted of a pre-heat of 95°C for 1 min, and 30 cycles of the following: 95°C for 45 s of denaturation, 50°C for 45 s of annealing, 72°C for 2 min of extension. A final extension of 72°C for 7 min was also included. The primers used to amplify the atpB-rbcL were from Samuel et al. (1997) while those for the rps16 were from Oxelmann et al. (1997). The protocols for atpB-rbcL and rps16 were similar to trnL-F but used a higher annealing temperature (52°C). Four primers were used to amplify the matK, they were: “19F” (Molvray et al. 2000), “9R” (Hilu et al. 1999), “390F” (Cuènoud et al. 2002), and “trnK2R” (Johnson and Soltis 1994). The PCR amplification protocol of matK consisted of a pre-heat of 94°C for 3 min, and 30 cycles of the following: 94°C for 1 min of denaturation, 52°C for 1 min of annealing, 72°C for 2.5 min of extension. A final extension of 72°C for 7 min was also employed. All successful PCR products were purified using the same procedure to tDNA purification but used ultra-pure sterile water instead of TE buffer as the elution buffer. DNA was sequenced using Applied Biosystems BigDye terminator kits, v.1.1, on an Applied Biosystems 310 automated DNA sequencer. The full sequences of all taxa listed in Table 2 were obtained. Only partial sequence of one taxon, Olyra latifolia, was used, because of difficulties with the matK 19F primer during sequencing.
DNA sequence editing, assembly, and phylogenetic analysis
DNA sequences were edited and assembled using AutoAssembler Software, version 2.1 (Applied Biosystems). The sequences were then imported to PAUP 4.0* Beta 2 (Swofford 1998) for alignment. Sequences were aligned by eye. Gaps were scored as additional binary characters (scoring gaps of identical size and position only). The resulting sequences were subjected to maximum-parsimony analysis using the heuristic search options in PAUP 4.0* Beta 2 (Swofford 1998). Searches included 1,000 replicates of random stepwise addition saving no more than 100 trees for tree bisection reconstruction branch swapping per replicate. Bootstrapping included 1,000 replicates and the same heuristic search settings as the individual searches, except that simple addition sequence was used instead of random stepwise addition.
Bayesian analyses were performed using MrBayes version 3.2 (Huelsenbeck and Ronquist 2001). The GTR + G model of substitutions was selected for each gene partition following hierarchical likelihood ratio tests (Huelsenbeck et al. 1996). Substitution rate matrix parameters and the shape of the Gamma distribution were estimated independently for each gene partition. The Markov chain Monte-Carlo (MCMC) algorithm was run for ten million generations over eight parallel chains, sampling every 1,000 generations on the Vital-it cluster of the Swiss Institute of Bioinformatics. Convergence of the MCMC was assessed using the Gelman and Rubin (1992) test as implemented in the R package Coda (Plummer et al. 2006) and the generations before convergence were discarded as burnin.
Results
Multi-gene region phylogenetic analysis
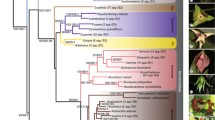
The matrix used for the multi-gene analysis was obtained from trnL-F, atpB-rbcL, rps16, and matK sequences. The sequences have been deposited in GenBank, EMBL, GDBJ under the accession numbers (EU434041–EU434104). The aligned matrix was 4,983 characters long, 3,326 were constant, 738 were variable but parsimony-uninformative, and 919 were parsimony informative. The tree search using maximum parsimony found three equally most parsimonious trees, of 2,688 steps (Fig. 1). CI and RI were 0.72 and 0.79, respectively. Bootstrap (BS) percentages (≥50%BS) are described as low (50–74%), moderate (75–84%), and high (85–100%). The Bayesian analysis produced the same topology as the parsimony analysis. Posterior probability (PP) values from the Bayesian analyses are provided in italics below the bootstrap value in Fig. 1.
One of three equally most parsimonious trees obtained from comparative sequence analysis of combined trnL-F, atpB-rbcL, rps16, and matK sequence data. Values above branches represent the number of steps supporting each branch. Values below branches represent the percentages of bootstrap supporting each branch. Numbers in italics represent the posterior probability value (PP). The BEP clade, the subfamial classification, and the tribal classification (first column on far right) are according to GPWG (2001). The geographical classification of woody bamboos (second column from far right) is shown according to Clark et al. (1995) and Ní Chonghaile (2002). The two subtribal classifications shown (the first and the second from far left) are according to Clayton and Renvoize (1986) and Ohrnberger (1999), respectively. N, neotropical woody bamboos; P, palaeotropical woody bamboos; T, temperate woody bamboos; ART, Arthrostylidiinae; ARU, Arundinariinae; BAM, Bambusinae; CHU, Chusqueinae; GUA, Guaduinae; HIC, Hickeliinae; MEL, Melocanninae; RAC, Racemobambosinae; SHI, Shibataeinae; THA, Thamnocalaminae
The BEP clade was highly supported (100%BS, 1.00PP) as monophyletic. The monophyly of each subfamily was also highly supported (100%BS, 1.00PP). Pooideae were sister to Bambusoideae s.s. with 99%BS (1.00PP) and Ehrhartoideae were sister to the lineage consisting of Pooideae and Bambusoideae s.s. (100%BS, 1.00PP).
The analysis showed that Olyreae (a monophyletic group with 100%BS, 1.00PP) were sister only to the tropical Bambuseae and not to the whole of Bambuseae. This suggests that Bambuseae are not monophyletic. There is 57%BS and 1.00PP for the sister-group status of Olyreae to the tropical Bambuseae. Division of the Bambuseae into temperate, neotropical, and palaeotropical groups was also evident. The temperate Bambuseae were highly supported (100%BS, 0.99PP) as monophyletic and were sister to the group consisting of Olyreae and the tropical woody bamboos. The tropical woody bamboos were also highly supported (91%BS, 1.00PP) as monophyletic, consisting of neotropical and the palaeotropical Bambuseae plus Mullerochloa moreheadiana from Australia and another species Neololeba atra from South Mindanao in the Philippines extending to Australia. The neotropical woody bamboos were moderately supported (77%BS, but had a PP value of 1.00) and the palaeotropical Bambuseae together with Mullerochloa moreheadiana and Neololeba atra were monophyletic with high support (91%BS, 1.00PP).
None of the subtribes of temperate woody bamboos, whether according to Clayton and Renvoize (1986) or Ohrnberger (1999), were monophyletic. However, several tropical subtribes according to Ohrnberger (1999) were monophyletic. Arthrostylidiinae, represented by Arthrostylidium and Rhipidocladum were monophyletic with high support (100%BS, 1.00PP). Guadua (Guaduinae) was sister to Arthrostylidiinae with high support (93%BS, 1.00PP). Chusquea (Chusqueinae) was sister to the group consisting of Arthrostylidiinae and Guaduinae (77%BS, 1.00PP). The analysis has also shown that subtribe Melocanninae (Schizostachydinae) was highly supported (100%BS, 1.00PP). Furthermore, Melocanninae were sister to the rest of the palaeotropical Bambuseae together with Mullerochloa moreheadiana and Neololeba atra, with high support (91%BS, 1.00PP). Temburongia represents subtribe Hickeliinae according to Ohrnberger (1999) and it was sister to the remaining palaeotropical woody bamboos plus Mullerochloa moreheadiana and Neololeba atra (99% BS, 1.00PP). The subtribe Bambusinae according to Ohrnberger (1999) was not monophyletic because the genus Vietnamosasa (representing Racemobambosinae of Ohrnberger 1999) was embedded within Bambusinae. Subtribe Bambusinae according to Clayton and Renvoize (1986) was also not monophyletic (Fig. 1).
Discussion
Phylogenetics of Bambusoideae s.s.
The subfamily Bambusoideae s.s., as defined by the GPWG (2001), was resolved as monophyletic, consisting of members from the herbaceous bamboos (tribe Olyreae) and a non-monophyletic assemblage of woody bamboos (tribe Bambuseae). The sister group to the Bambusoideae s.s. was Pooideae. This confirms the monophyly of Bambusoideae s.s. as found in previous analyses (Clark et al. 1995; GPWG 2001; Bouchenak-Khelladi et al. 2008). The secondary gain of the pseudopetiole and the secondary loss of the lamina of the first seedling leaf were recognized by the GPWG (2001) as synapomorphies for Bambusoideae s.s. The presence of strongly asymmetrically invaginated arm cells, as suggested by Zhang and Clark (2000), may also be a potential synapomorphy. The natural distribution of Bambusoideae s.s. is wide, ranging approximately from 46°N to 47°S latitude and from sea level to as much as 4,300 m in equatorial highlands (Judziewicz et al. 1999).
One tribe of Bambusoideae s.s., Olyreae, was supported as monophyletic with high bootstrap support. However, the combined analysis failed to support the monophyly of the other bambusoid tribe, Bambuseae. Bambuseae were paraphyletic and can be divided into two clades based on their molecular variation and geographical distribution. The tropical woody bamboos were sister to Olyreae (57%BS, 1.00PP, Fig. 1) while the temperate woody bamboos were sister to the group consisting of Olyreae and tropical woody bamboos (100%BS, 1.00PP). The non-monophyly of Bambuseae was also found in a combined analysis of taxa spanning the whole of the grass family (Bouchenak-Khelladi et al. 2008). This combined study, those of Bouchenak-Khelladi et al. (2008), and the Bamboo Phylogeny Group (BPG; recent personal communication) showed that Olyreae are sister to the tropical Bambuseae. There is, therefore, strong evidence that Bambuseae (sensu Clayton and Renvoize 1986; Ohrnberger 1999) are not monophyletic and that Olyreae are the sister group of the tropical Bambuseae. Thus, the temperate woody bamboos are best accommodated at tribal level as Arundinarieae Nees ex Ascherson and Graebner (name validated in 1902). Under this new scenario the Bambusoideae s.s. should include three tribes: Olyreae, Bambuseae s.s., and Arundinarieae (Fig. 2). In terms of character evolution, there are two equally parsimonious scenarios for the evolution of woodiness, one involving parallel evolution and the other reversal. Either the Bambuseae s.s. and Arundinarieae independently evolved woodiness from a herbaceous ancestral state or, alternatively, the ancestor of the Bambuseae s.s., Arundinarieae, and Olyreae group was woody and Olyreae subsequently reversed to a herbaceous state of their non-bambusoid sister group. We can only speculate which of these two scenarios is the most likely. Woodiness has evolved a number of times independently in the grasses, for example in some Panicoideae and Arundinoideae. It is therefore not unlikely that there has been parallel gain of woodiness in Bambusoideae s.s.
Cladogram showing relationships within Bambusoideae s.s. and other grasses. The grey box indicates the BEP clade (Bambusoideae, Ehrhartoideae, Pooideae) according to GPWG (2001). The tribal classification (Arundinarieae, Bambuseae s.s., and Olyreae) is according to this study where Arundinarieae and Bambuseae s.s. are redefined whereas Olyeae (herbaceous bamboos) is as defined by GPWG (2001). The geographical classification of woody bamboos is shown according to Clark et al. (1995) and Ní Chonghaile (2002). M, Melocanninae according to Ohrnberger (1999) is sister to rP&A (= the rest of palaeotropical plus Austral woody bamboo subtribes). For the distribution of Olyreae, the lighter shade of black colour indicates uncertainty about whether it is truly native to these areas. The distribution of Arundinarieae found in the tropical zone is from the high elevation, usually from 1,000 m to as high as 3,630 m (adapted from Ohrnberger 1999). All the distribution maps were adapted from http://www.eeob.iastate.edu/research/bamboo/maps.html, with permission from Dr Lynn Clark
The geographical division of woody bamboos into temperate, palaeotropical, and neotropical groups (Clark et al. 1995) could be generally applied to our results. The temperate woody bamboos were highly supported. Palaeotropical woody bamboos plus Mullerochloa moreheadiana and Neololeba atra (palaeotropical and Austral woody bamboos), were supported (91%BS, 1.00PP, Fig. 1). This is the first time an Australian Bambuseae genus, Mullerochloa, and a genus extending to Australia, Nelololeba, have been included in phylogenetic analyses. The topology of having Neololeba sister to Dinochloa and Mullerochloa is a novel result. Temburongia is a monotypic genus (T. simplex) from Brunei (Dransfield and Wong 1996), representing subtribe Hickeliinae according to Ohrnberger (1999). It was the most outlying group within the palaeotropical and Austral woody bamboos. Thus a lineage represented by Temburongia, a lineage represented by a group consisting of Neololeba and Dinochloa, and another lineage represented by Mullerochloa are successively sister to the rest of the palaeotropical and Austral Bambuseae s.s. The neotropical woody bamboos were also resolved as monophyletic (77%BS, 1.00PP). The previous phylogenetic analyses of this group (e.g. Clark et al. 1995; Kelchner and Clark 1997; Zhang 2000) are congruent with our study. The temperate woody bamboos (here recognized as Arundinarieae) were supported as monophyletic, but none of their subtribes according to Clayton and Renvoize (1986) or Ohrnberger (1999) were supported. A limited amount of supported resolution was found within Arundinarieae in our analyses. For example, Borinda, Chimonobambusa, and Menstruocalamus group together (62%BS, 0.99PP).
Neotropical woody bamboos
A group containing Arthrostylidiinae, Chusqueinae, and Guaduinae sensu Ohrnberger (1999) were resolved. Arthrostylidiinae, represented by Arthrostylidium and Rhipidocladum, were positioned in Bambusinae by Clayton and Renvoize (1986), and were highly supported in our analysis (100%BS, 1.00PP). Guadua, representing Guaduinae, was sister to Arthrostylidiinae (93%BS, 1.00PP). Guadua was treated under Bambusa in Bambusinae by Clayton and Renvoize (1986). There was no evidence from the results of single or combined analyses to support this placement. Other molecular studies, that sampled Guaduinae according to Ohrnberger (1999), have shown that they were also sister to Arthrostylidiinae (Kelchner and Clark 1997; Zhang 2000; Bouchenak-Khelladi et al. 2008). Chusqueinae, represented by Chusquea, were sister to the group consisting of Arthrostylidiinae and Guaduinae (77%BS, 1.00PP). This relationship is generally congruent with previous studies (Kelchner and Clark 1997; Zhang 2000; Bouchenak-Khelladi et al. 2008). All three of these subtribes are from Central and South America (Ohrnberger 1999).
Palaeotropical and Austral woody bamboos
Subtribe Melocanninae (Schizostachydinae) according to Ohrnberger (1999) was well supported and Melocanninae were sister to the rest of the palaeotropical and Austral woody bamboos (91%, 1.00PP). This relationship is a novel result. Melocanninae are generally found at lower elevations in South, South-East, and East Asia (Table 1). Morphologically, this subtribe has pseudo-spikelets with a distinctive glabrous ovary that bears an elongated and persistent style usually divided into three short stigmas (Soderstrom and Ellis 1987). Anatomically, they differ from other subtribes in having larger microhairs, in the presence of refractive papillae, and a pronounced S-shaped keel with complex vasculature in its leaf-blade (Soderstrom and Ellis 1987). These are synapomorphic characters that set Melocanninae apart from the rest of the palaeotropical and Austral woody bamboos. Genus Pseudostachyum was placed under Schizostachyum by Clayton and Renvoize (1986). However, our analyses showed that Pseudostachyum was sister to the remaining Melocanninae (100%BS, 1.00PP) and support the generic status of Pseudostachyum. This finding is congruent with the classifications adopted by Soderstrom and Ellis (1987), Dransfield and Widjaja (1995), Clark (1995), Li (1998) and Ohrnberger (1999). Cephalostachyum, Neohouzeaua and Schizostachyum, were monophyletic (99%BS, 1.00PP) within Melocannineae. However, the relationships among these genera were unclear. Soderstrom and Ellis (1987) placed Neohouzeaua under Schizostachyum. Clayton and Renvoize (1986) treated Neohouzeaua and Cephalostachyum as synonymous with Schizostachyum, whereas Dransfield and Widjaja (1995), Clark (1995), Li (1998), and Ohrnberger (1999) treated them as separate genera. Our results are more consistent with the Schizostachyum s.l. hypothesis of Clayton and Renvoize (1986). The recently established genus Temburongia, from Ulu Temburong National Park, Brunei (Dransfield and Wong 1996), was the only representative of the subtribe Hickeliinae according to Ohrnberger (1999) in our analyses. Temburongia was treated as incertae sedis by Clark et al. (2007). In our study it was sister to the remaining palaeotropical and Austral woody bamboos (99%BS, 1.00PP).
Subtribe Bambusinae
None of our analyses supported the monophyly of subtribe Bambusinae according to Ohrnberger (1999), because Vietnamosasa, which he positioned in Racemobambosinae, was embedded within it. All analyses also showed that Bambusinae according to Clayton and Renvoize (1986) are polyphyletic, because its taxa were distributed across the phylogenetic tree of Bambuseae. Like Racemobambos, Vietnamosasa has a determinate inflorescence (Dransfield 2000a) instead of an indeterminate inflorescence as found in Bambusinae. However, if Vietnamosasa (and Temochloa, also possessing a determinate inflorescence, Dransfield 2000b) could be included within Bambusinae, then we show no evidence against the monophyly of Bambusinae sensu Ohrnberger (1999).
There are four recently established genera (Mullerochloa, Neololeba, Phuphanochloa, and Temochloa) that can be included within Bambusinae according to Ohrnberger (1999). Phuphanochloa is a new genus from northeastern Thailand, composed of a single species, P. speciosa (Sungkaew et al., in press). It is morphologically similar to, and phylogenetically related to, Bambusa, Dendrocalamus, and Gigantochloa (Bambusinae). The results showed that Phuphanochloa is sister to a group consisting of Bambusa beecheyana, B. malingensis, B. oldhamii, and Neosinocalamus affinis (= B. affinis). The other newly established genera, Mullerochloa, Neololeba, and Temochloa, may be best interpreted as phylogenetically outlying within a broadly circumscribed Bambusinae or they could merit subtribe status (Fig. 1). Temochloa is a monotypic and endemic genus (T. liliana) from limestone regions of southern Thailand, which has no subtribe applied to it (Dransfield 2000b). Mullerochloa is a monotypic and endemic genus (M. moreheadiana) from Queensland, Australia (Wong 2005). Neololeba (represented by N. atra, see Table 2) is a relatively new genus established to accommodate five bamboos known from South Mindanao in the Philippines, North Sulawesi, Moluccas, New Guinea, Solomon Islands, and Queensland, Australia (Widjaja 1997). Ohrnberger (1999), without referring to Widjaja (1997), treated this species under Bambusa as B. atra Lindley and placed it in Bambusinae. A Neololeba and Dinochloa group was sister to the remaining palaeotropical and Austral woody bamboos including Temochloa and Mullerochloa (61%BS, 0.97PP). Dinochloa is mainly distributed on the Malay Peninsula, in Borneo, Indonesia, and Philippines, extending to southern Thailand and the Andaman and Nicobar Islands (Dransfield 1981).
Dinochloa and Melocalamus are consistently classified in Bambusinae (Clayton and Renvoize 1986; Soderstrom and Ellis 1987; Dransfield and Widjaja 1995; Clark 1995; Li 1998; Ohrnberger 1999). Surprisingly, Dinochloa malayana was sister to Neololeba atra (100%BS, 1.00PP). On the basis of morphology we might expect Dinochloa to group either with Melocalamus or Mullerochloa, rather than with Neololeba (McClure 1966; Dransfield 1981; Wong 1995; Li and Stapleton 2006). This suggests that one reproductive character (berrylike caryopsis with thick and fleshy pericarp) and two vegetative characters (presence of an abrupt swelling of the very basal part of the culm internodes and presence of a rugose basal zone of the culm sheath) are homoplasious because they have evolved independently among these three genera.
Apart from the most outlying taxon, Temburongia, the biogeographical range of the remaining outlying lineages (Neololeba/Dinochloa and Mullerochloa) is outside that of the core Bambusinae. This suggests that the ancestors of Bambusinae were from somewhere in mainland Asia, possibly south China, India, or even mainland Southeast Asia and two outlying lineages evolved separately from southern Thailand and Malaysia to northern Australia. The paucity of bamboo species in Australia would also support this hypothesis. If the origin of these bamboos had been near Australia we would expect much higher diversification in this area.
Oreobambos and Oxytenanthera were sister taxa (100%BS, 1.00PP). Both Oreobambos and Oxytenanthera are monotypic genera, naturally distributed in tropical Africa (Ohrnberger 1999). Clayton and Renvoize (1986) treated these two genera in different subtribes (Fig. 1). However, it is geographically (Ohrnberger 1999) and phylogenetically (this study) clear that they should be grouped together and included in Bambusinae.
Morphologically, the genus Dendrocalamus is similar to several other genera including Bambusa, Dendrocalamopsis, Gigantochloa, Houzeaubambus, Klemachloa, Oreobambos, Oxytenanthera, Sinocalamus, and Neosinocalamus (Holttum 1958; McClure 1966; Clayton and Renvoize 1986; Soderstrom and Ellis 1987; Dransfield and Widjaja 1995; Wong 1995; Stapleton and Xia 1997; Li 1997; Li and Xue 1997; Li 1998; Ohrnberger 1999; Li and Stapleton 2006) and our results confirmed that Dendrocalamus is closely related to Bambusa, Dendrocalamopsis (= Bambusa), and Gigantochloa. Our results also showed that Melocalamus grouped with Dendrocalamus (100%BS, 1.00PP, Fig. 1). This relationship has never been reported before. However, it could be because of hybridization (S. Sungkaew et al., in preparation). Melocalamus was expected to group with Dinochloa or Mullerochloa on the basis of morphology. The results also showed that Bambusa oldhamii groups with Bambusa beecheyana, Bambusa malingensis, and Neosinocalamus affinis (= Bambusa emeiensis), with 80%BS (1.00PP). This would be consistent with the placement of Bambusa oldhamii in Bambusa. The delimitation of Sinocalamus has proven extremely difficult (McClure 1940; Raizada 1948; Chia and Fung 1980; Xia and Stapleton 1997). Our results do not support the recognition of Sinocalamus (represented by S. oldhamii, see Table 2) and indicate that its species are better placed in Bambusa than Dendrocalamus.
To conclude, we have conducted one of the most comprehensive multi-gene region phylogenetic studies on Bambusoideae s.s. by including over 60 taxa representing all the subtribes of the traditionally recognized tribe Bambuseae and a representative sample of related taxa including Olyreae. The results have resolved a number of patterns, summarized in Fig. 2, that were previously unrecognized or poorly supported, for example:
-
1
the sister-group status of Pooideae to Bambusoideae s.s.;
-
2
the non-monophyly of Bambuseae;
-
3
the sister-group status of Olyreae to the tropical Bambuseae;
-
4
the sister-group status of temperate Bambuseae to a tropical Bambuseae/Olyreae clade;
-
5
the sister-group status of Melocanninae to the remaining palaeotropical and Austral Bambuseae; and
-
6
the division of Bambuseae s.s. into Neotropical and Palaeotropical/Austral groups.
The results indicate a need to revise the classification of Bambuseae and we recommend use of the tribal name Arundinarieae to accommodate the temperate woody bamboos.
References
Andersson L, Chase MW (2001) Phylogeny and classification of Marantaceae. Bot J Linn Soc 135:275–287
Ascherson P, Graebner P (1902) Synopsis der mitteleuropäischen Flora. Leipzig 2:705–795
Asmussen CB, Chase MW, Baker WJ, Dransfield J (2000) Phylogeny of the palm family (Arecaceae) based on rps16 intron and trnL-trnF plastid DNA sequences. In: Wilson KL, Morrison DA (eds) Monocots: systematics and evolution. CSIRO, Collingwood, pp 525–535
Bouchenak-Khelladi Y, Salamin N, Savolainen V, Forest F, van der Bank M, Chase MW, Hodkinson TR (2008) Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol Phylogenet Evol 47:488–505
Chase MW, Hills HH (1991) Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40:215–220
Chia LC, Fung HL (1980) On the validity of the genera Sinocalamus McClure and Lingnania McClure. Acta Phytotaxon Sin 18:211–216
Clark LG (1995) Bamboo systematics today. Eur Bamboo Soc J 6:40–46
Clark LG, Zhang W, Wendel JF (1995) A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Syst Bot 20:436–460
Clark LG, Dransfield S, Triplett JK, Sánchez-Ken JG (2007) Phylogenetic relationships among the one-flowered, determinate genera of Bambuseae (Poaceae: Bambusoideae). Aliso 23:315–332
Clayton WD, Renvoize SA (1986) Genera Graminum, grasses of the world. Kew Bull Addit Ser XIII. Her Majesty's Stationery Office, London
Clayton WD, Harmann KT, Williamson H (2008) GrassBase—the online world grass flora. Royal Botanic Gardens, Kew. Online at http://www.rbgkew.org.uk/data/grasses-db/. Accessed 27 Sep 2008
Cuènoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89:132–144
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull Bot Soc Am 19:11–15
Dransfield S (1981) The genus Dinochloa (Gramineae-Bambusoideae) in Sabah. Kew Bull 36:613–633
Dransfield S (2000a) Notes on ‘Pek’ and ‘Chote’, members of the genus Vietnamosasa (Poaceae-Bambusoideae) in Thailand. Thai Forest Bull, Bot 28:163–176
Dransfield S (2000b) Temochloa, a new bamboo genus (Poaceae-Bambusoideae) from Thailand. Thai Forest Bull, Bot 28:179–182
Dransfield S, Widjaja EA (1995) Plant resources of South-East Asia No. 7: Bamboos. Backhuys, Leiden
Dransfield S, Wong KM (1996) Temburongia, a new genus of bamboo (Gramineae: Bambusoideae) from Brunei. Sandakania 7:49–58
Friar E, Kochert G (1994) A study of genetic variation and evolution of Phyllostachys (Bambusoideae: Poaceae) using nuclear restriction fragment length polymorphisms. Theor Appl Genet 89:265–270
Gelman A, Rubin D (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–511
Gielis J, Everaert I, De Loose M (1997) Genetic variability and relationships in Phyllostachys using random amplified polymorphic DNA. In: Chapman GP (ed) The bamboos. Linn Soc Symp Ser 19:107–124
GPWG (Grass Phylogeny Working Group) (2001) Phylogeny and subfamily classification of the grasses (Poaceae). Ann Mo Bot Gard 88(3):373–430
Hilu KW, Alice LA, Liang HP (1999) Phylogeny of Poaceae inferred from matK sequences. Ann Mo Bot Gard 86:835–851
Hodkinson TR, Renvoize SA, Ní Chonghaile G, Stapleton CMA, Chase MW (2000) A comparison of ITS nuclear rDNA sequence data and AFLP markers for phylogenetic studies in Phyllostachys (Bambusoideae, Poaceae). J Plant Res 113:259–269
Hodkinson TR, Chase MW, Lledo D, Salamin N, Renvoize SA (2002) Molecular phylogeny of Miscanthus s.l., Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) using DNA sequences from the ITS nuclear ribosomal DNA and the plastid trnL-F regions. J Plant Res 115:381–392
Hodkinson TR, Salamin N, Chase MW, Bouchenak-Khelladi Y, Renvoize SA, Savolainen V (2007a) Large trees, supertrees, and diversification of the grass family. Aliso 23:248–258
Hodkinson TR, Waldren S, Parnell JAN, Kelleher CT, Salamin K, Salamin N (2007b) DNA banking for plant breeding, biotechnology and biodiversity evaluation. J Plant Res 120:17–29
Holttum RE (1958) The bamboos of the Malay Peninsula. Gard Bull Singapore 16:1–135
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Huelsenbeck JP, Bull JJ, Cunningham CW (1996) Combining data in phylogenetic analysis. Trends Ecol Evol 11:152–158
Johnson LA, Soltis DE (1994) matK DNA sequences and phylogenetic reconstruction in Saxifragaceae sensu stricto. Syst Bot 19:143–156
Judziewicz EJ, Clark LG, Londoño X, Stern MJ (1999) American bamboos. Smithsonian Institution Press, Washington and London
Kelchner SA, Clark LG (1997) Molecular evolution and phylogenetic utility of the chloroplast rpl16 intron in Chusquea and the Bambusoideae (Poaceae). Mol Phylogenet Evol 8:385–397
Li DZ (1997) The flora of China Bambusoideae Project-problems and current understanding of bamboo taxonomy in China. In: Chapman GP (ed) The bamboos, vol. 19. Linnean Society Symposium Series, pp 61–81
Li DZ (1998) Taxonomy and biogeography of the Bambuseae (Gramineae: Bambusoideae). In: Rao AN, Rao VR (eds) Proceedings of training course/workshop 10–17 May 1998, Kunming and Xishuanbanna, Yunnan, China, pp 14–23
Li DZ, Stapleton CMA (2006) Melocalamus Bentham. Fl China 22:48–49
Li DZ, Xue JR (1997) The biodiversity and conservation of bamboos in Yunnan, China. In: Chapman GP (ed) The bamboos, vol. 19. Linnean Society Symposium Series, pp 83–94
Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, A SCM (2006) Tribe Bambuseae. Fl China 22:11–180
Liang H, Hilu KW (1996) Application of the matK gene sequences to grass systematics. Can J Bot 74:125–134
McClure FA (1940) New genera and species of Bambusaceae from eastern Asia. Lingnan Univ Sci Bull 9:66–67
McClure FA (1966) The bamboos: a fresh perspective. Harvard University Press, Cambridge
Molvray M, Kores PJ, Chase MW (2000) Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA (eds) Monocots: systematics and evolution. CSIRO, Collingwood, pp 441–448
Ní Chonghaile G (2002) Molecular systematics of the woody bamboos (Bambuseae). Ph.D. thesis, University of Dublin, Trinity College, Dublin
Ohrnberger D (1999) The bamboos of the world: annotated nomenclature and literature of the species and the higher and lower taxa. Elsevier Science, Amsterdam
Oxelmann B, Liden M, Berglund D (1997) Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst Evol 206:257–271
Plummer M, Best N, Cowles K, Vines K (2006) CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11
Raizada MB (1948) A little-known Burmese bamboo. Indian For 74:7–10
Reeves G, Chase MW, Goldblatt P, Rudall P, Fay MF, Cox AV, Lejeune B, Sousa-Chies T (2001) Molecular systematics of Iridaceae: evidence from four plastid DNA regions. Am J Bot 88:2074–2087
Salamin N, Hodkinson TR, Savolainen V (2002) Building supertrees: an empirical assessment using the grass family (Poaceae). Syst Biol 51:136–150
Samuel R, Pinsker W, Kiehn M (1997) Phylogeny of some species of Cyrtandra (Gesneriaceae) inferred from the atpB/rbcL cpDNA intergene region. Bot Acta 110:503–510
Soderstrom TR, Ellis RP (1987) The position of bamboo genera and allies in a system of grass classification. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution Press, Washington, D.C., pp 225–238
Soltis DE, Soltis PS (1998) Choosing and approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematics of plants II, DNA sequencing. Kluwer Academic, Dordrecht, pp 1–41
Stapleton CMA (1997) The morphology of woody bamboos. In: Chapman GP (ed) The bamboos, vol. 19. Linnean Society Symposium Series, pp 251–267
Stapleton CMA, Xia NH (1997) A new combination in Bambusa (Gramineae-Bambusoideae). Kew Bull 52:235–238
Sun Y, Xia NH, Lin R (2005) Phylogenetic analysis of Bambusa (Poaceae: Bambusoideae) based on Internal Transcribed Spacer sequences of nuclear ribosomal DNA. Biochem Genet 43:603–612
Sungkaew S, Teerawatananon A, Parnell JAN, Stapleton CMA, Hodkinson TR (in press) Phuphanochloa, a new bamboo genus (Poaceae: Bambusoideae) from Thailand. Kew Bull
Swofford DL (1998) Phylogenetic analysis using Parsimony (PAUP) version 4.0. Sinauer Associates, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Watanabe M, Ito M, Kurita S (1994) Chloroplast DNA phylogeny of Asian Bamboos (Bambusoideae, Poaceae) and its systematic implication. J Plant Res 107:253–261
Widjaja EA (1997) New taxa in Indonesian bamboos. Reinwardtia 11:57–152
Wong KM (1995) The bamboos of Peninsular Malaysia. Malayan Forest Records No. 41, Forest Research Institute Malaysia (FRIM), Kuala Lumpur, Malaysia
Wong KM (2005) Mullerochloa, a new genus of bamboo (Poaceae: Bambusoideae) from north-east Australia and notes on the circumscription of Bambusa. Blumea 50:425–441
Xia NH, Stapleton CMA (1997) A new combination in Dendrocalamus (Gramineae: Bambusoideae). Kew Bull 52:483–485
Yang HQ, Peng S, Li DZ (2007) Generic delimitations of Schizostachyum and its allies (Gramineae: Bambusoideae) inferred from GBSSI and trnL-F sequence phylogenies. Taxon 56:45–54
Zhang W (2000) Phylogeny of the grass family (Poaceae) from rpl16 intron sequence data. Mol Phylogenet Evol 15:135–146
Zhang W, Clark LG (2000) Phylogeny of classification of the Bambusoideae (Poaceae). In: Jacobs SWL, Everett JE (eds) Grasses: systematics and evolution. CSIRO, Collingwood, pp 35–42
Acknowledgements
We thank several people who helped or provided us with the plant material used in this study: Dr Soejatmi Dransfield, Dr Wang Hong, Mr Gareth Hodkinson, Dr Surrey Jacobs, Dr Wong Khoon Meng, Dr Ruth Kiew, Dr Duangchai Sookchaloem, and Ms Atchara Teerawatananon. We are grateful to Dr Soejatmi Dransfield who has contributed significantly to this paper. Special thanks to Dr Vincent Savolainen, Dr Mark Chase, and Mr Laszlo Csiba who helped with the molecular work. This work was supported by: the TRF/BIOTEC Special Program for Biodiversity Research and Training grant T_147003; a Trinity College Dublin, Eire, Postgraduate Studentship and the Trinity College Postgraduate Travel Reimbursement Fund; and the Faculty of Forestry, Kasetsart University, Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sungkaew, S., Stapleton, C.M.A., Salamin, N. et al. Non-monophyly of the woody bamboos (Bambuseae; Poaceae): a multi-gene region phylogenetic analysis of Bambusoideae s.s.. J Plant Res 122, 95–108 (2009). https://doi.org/10.1007/s10265-008-0192-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-008-0192-6