Abstract
Sonchus gandogeri, a woody sow-thistle, is an endangered Canary Island endemic with only two known populations, one in the El Golfo and another in the Las Esperillas of El Hierro. Amplified fragment length polymorphism (AFLP) markers were used to assess the genetic variation within and among populations. The mean genetic diversity of two populations was estimated to be 0.380, and the El Golfo population (0.380) had higher genetic diversity than the southeastern one (0.268). The unbiased Nei’s genetic identity between the two populations was 0.846. The mean genetic diversity of S. gandogeri was much higher than that of the other endangered plant species. This is perhaps due to breeding system, life form, extinction, and/or introgressive hybridization and hybrid origin of the taxon. This study also indicates that the two populations are not strongly differentiated (GST=0.149). This study suggests that S. gandogeri is more likely to become extinct due to environmental or demographic forces than genetic factors, such as inbreeding depression. More strict control of introduced herbivores is necessary to protect these populations, and germplasm collection for ex situ conservation is needed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic variation within a taxon is thought to be crucial for the long-term survival and continued evolution of populations or species (Franklin 1980; Beardmore 1983; Frankel 1983; Huenneke 1991). Thus, an accurate estimate of the level and distribution of genetic diversity of threatened and endangered species is an important element in designing conservation programs (Hamrick et al. 1991; Schaal et al. 1991; Chalmers et al. 1992; Cardoso et al. 1998). Studies on genetic diversity within and among populations of insular endemic plant species using molecular markers have increased in recent years due to their central importance in planning in situ and ex situ conservation efforts (e.g., de Joode and Wendel 1992; Gemmill et al. 1998; Francisco-Ortega et al. 2000; Crawford et al. 2001; Batista et al. 2001; Bouza et al. 2002). Three reviews compiled from both the Pacific and Atlantic Oceans (de Joode and Wendel 1992; Francisco-Ortega et al. 2000; Crawford et al. 2001) showed low diversity in insular endemics in general and suggested that there is lower variation within species of Pacific Island endemics than in the Canary Island endemics. These reviews also found a high proportion of diversity within populations.
The woody Sonchus alliance (Asteraceae: Sonchinae) is endemic to the Macaronesia islands (with the exception of one species, S. pinnatifidus Cav., occurring both in the Canaries and Morocco) and is comprised of six genera and approximately 31 species (Kim et al. 1996a, 1996b). The alliance has been regarded as an outstanding example of adaptive radiation among angiosperms in Macaronesia (Aldridge 1975, 1979). Recent molecular phylogenetic and isozyme studies support the alliance’s single origin and recent radiation on the Macaronesian islands (Kim et al. 1996a, 1996b, 1999).
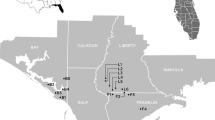
Sonchus gandogeri Pitard, one of two endangered Sonchus species, is a shrub up to 1.8 m tall and is endemic to the island of El Hierro, the youngest (0.8 Ma old) and western most island (Fig.1). The leaves are coriaceous, somewhat glossy, and pinnatisect with long, slender lobes about 5–10 mm wide (Boulos 1974; Bramwell and Bramwell 2001). The inflorescence is a corymb of about 20–60 heads, with about 60 florets per head and a diameter of 1–2 cm. This species is very rare and sporadic on the southeastern cliffs of the island as well as in the El Golfo region on the northeastern cliffs of Frontera, Las Castitas, and Los Llanillos (Fig.1b; Bramwell and Bramwell 2001).
a Five Macaronesian archipelagos located in the northeast Atlantic Ocean and the Canary archipelago and the age of the islands in Ma (shown in parentheses) (Carracedo 1994). b The map of El Hierro, showing the location of two Sonchus gandogeri populations. Contour intervals are 400 m
We have observed only two recognizable Sonchus gandogeri populations in El Hierro: one on the north-facing cliffs between Guinea and Las Puntas in the northeastern El Golfo region and another on the southeastern-facing cliffs near Las Esperillas (Fig.1b). These two populations occur at relatively lower elevations (<300 m) and are geographically isolated from one another; they are separated by a mountain range with an altitude above 1,000 m and a direct map distance of ca. 15 km. Although the El Golfo population is located on the north-facing cliffs, it is not under the direct influence of northeastern trade winds due to protection by the big cliffs. Furthermore, this population tends to occur in the northwestern-facing dry vertical cliffs because of exposure to the afternoon sun (A. Santos-Guerra, personal observation). In contrast, the southeastern population near Las Esperillas tends to occur on the lower east-facing cliffs, exposed to the northeastern trade winds and shaded in the afternoon by the cliffs. There has been no demographic survey of this taxon in either locality, and thus we do not know the exact number of individuals for either population. However, populations located in both regions are similar in size (approximately 50 individuals in each location).
Despite the rarity and endangered status of Sonchus gandogeri, there is currently no population genetic study and conservation management plan for this species. As an initial step in developing such a plan, we have assessed the genetic diversity and population structure of two natural populations of S. gandogeri using amplified fragment length polymorphisms (AFLPs) (Vos et al. 1995). The AFLP technique has been used successfully in plant population genetic studies, especially for endangered species (e.g., Travis et al. 1996; Palacios et al. 1999; Drummond et al. 2000; Schmidt and Jenssen 2000).
We have tested the following two hypotheses: (1) The two populations have low genetic diversity due to recent speciation and small population size, and (2) the two populations show significant genetic differentiation because of physical isolation and genetic drift.
Methods
Plant material
Young leaf tissue was collected from 22 individuals in each of the El Golfo and Las Esperillas populations and dried in silica gel. The sample sizes were small because many individuals were inaccessible due to their occurrence in vertical cliffs. All samples represent pure S. gandogeri based on their morphological characteristics and include both young and mature individuals at least several meters apart.
DNA extraction and AFLP reaction
Total genomic DNA was isolated from 25 mg of dried leaf tissue using DNeasy plant mini kits (QIAGEN, Chatsworth, CA, USA). The AFLP procedure was the same as that described by Vos et al. (1995) with minor modifications (Kim and Rieseberg 1999). Genomic DNA (300 ng) was digested for 1 h at 37°C with three units of EcoRI and MseI, 4 mg bovine serum albumin (BSA), 4 μl 10× buffer 2 (New England Biolabs, Beverly, MA, USA), and ddH2O to a final volume of 40 μl. Adapters were ligated to the digested fragments by adding 15 pmol EcoRI adapters, 150 pmol MseI adapters, 0.5 μl T4 polynucleotide DNA ligase, 1 μl 10 nm ATP, 1 mg BSA, 2.0 μl 10× buffer 2, and ddH2O, to a total volume of 50 μl and incubated for 3 h at 37°C.
Preamplification reactions were performed with two AFLP primers having a single selective nucelotide (A). The preamplification reactions contained 1 μl of template DNA from the ligation reaction, 187.5 ng EcoRI+A and 187.5 ng MseI+A primers, 0.4 μl dNTPs (each at 25 nm), 2.5 μl PCR buffer (500 mM KCl, 15 nm MgCl2, and 200 nm Tris–HCl), 0.4 units Taq polymerase, and ddH2O in a total volume of 25 μl. The reactions were placed in a thermal cycler (PTC-100, MJ Research, Watertown, MA, USA) programmed for 20 cycles, each consisting of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C.
Once the preamplifications were complete, selective amplifications were performed using 2.5 μl of 1:20 diluted preamplification reaction as a template, 5 ng of the EcoRI+3 nucleotide selective primer (Vos et al. 1995), 15 ng of MseI+3 nucleotide selective primer, 0.16 μl dNTPs, 1 μl PCR buffer, Taq polymerase, and ddH2O to a final volume of 10 μl. Amplifications were conducted in a MJ Research thermal cycler programmed for 36 cycles, each consisting of 30 s at 94°C, 90 s at 65°C (see below), and 90 s at 72°C. The 65°C annealing temperature of the first cycle was subsequently reduced by 1°C for the next 10 cycles and then continued at 54°C for the remaining 26 cycles. Likewise, the extension time of 90 s was reduced to 1 min for the last 26 cycles. All enzymes and buffers were purchased from New England Biolabs (Beverly, MA, USA). Adapters and primer sequences are the same as those described in Vos et al. (1995). The EcoRI+3 primers were labeled with IRD 700 and IRD 800 fluorescence dyes (Li-Cor, Lincoln, NE, USA).
Following amplification, reaction products were mixed with an equal volume (10 μl) of formamide dye (98% formamide, 10 mM EDTA pH 8.0, and bromphenol blue). The resulting mixtures were heated for 3 min at 90°C, quickly cooled on ice, and 1.2 μl was immediately loaded on KBplus 6.5% Gel Matrix (Li-Cor, Lincoln, NE, USA) polyacrylamide gels. The 50–700 size standard (Li-Cor, Lincoln, NE, USA) was run with the samples to estimate the size of fragments. Electrophoresis was performed at constant voltage (1,500 V) for 3 h at 43°C using an automated DNA sequencer (Li-Cor IR2, Nebraska, NE, USA).
Data analysis
The AFLP bands were scored manually as either presence or absence of each fragment within each individual and pooled over all fragments and primer combinations. Analysis was restricted to the 143 loci with reliably scored bands and a frequency of less than 95% for the most common phenotype. We determined the approximate sizes of fragments on the basis of a 50–700 size standard.
Based on the assumption that each locus is a two-allele system in Hardy-Weinberg equilibrium, we used PopGene 1.31 (Yeh et al. 1997) to calculate Nei’s (1973) gene diversity statistics (HT, HS, and GST) as well as the effective number of migrants per generation (Nm=0.5(1−GST)/GST), which is an indirect estimate of gene flow between the two populations. We also treated AFLP patterns as restriction fragment presence/absence data (RFLP) and used Nei and Li’s method (1979) using PAUP (Version 4b10, Swofford 2000) to calculate pairwise distances. This pairwise matrix was then used to construct a dendrogram by the neighbor-joining method (Saitou and Nei 1987).
Results
We surveyed 32 primer combinations, EcoRI+3 and MseI+3 selective primer pairs, and 12 of them showed banding patterns with very high reproducibility and clear band resolution. Polymorphic marker per primer combination ranged from 5 (E-AGC/M-AGA) to 28 (E-ACG/M-AGA).
The mean diversity value of the El Golfo population was 0.380 (±0.14, SD), whereas that of the Las Esperillas population is 0.268±0.18. The Ewens-Watterson test for neutrality (Manly 1985) indicated that the majority of AFLP loci, i.e., 93%, were neutral. The Nei’s genetic diversity for two populations of Sonchus gandogeri was 0.380±0.11.
Total genetic diversity (HT) within Sonchus gandogeri was 0.380±0.01, while the diversity within populations (HS) was 0.323±0.01 (Table 1). GST, the proportion of total diversity among populations, was 0.149 and Nm, the estimate of gene flow from GST, was 2.856. Nei’s (1978) unbiased measures of genetic identity and genetic distance between two populations were 0.846 and 0.168, respectively.
An unrooted neighbor-joining tree based on Nei and Li (1979) distance is shown in Fig. 2. The dendrogram shows that the Las Esperillas individuals fall into a single clade. The El Golfo population is paraphyletic, with some individuals forming a basal clade in the tree, but the majority of the individuals forming a large separate clade that falls within the larger clade containing the Las Esperillas individuals.
Discussion
Genetic diversity within populations and within a species
Overall, it appears that total genetic diversity in Sonchus gandogeri is much higher than that of other endangered species studied using AFLPs. Genetic diversity estimates in other AFLP studies have ranged only from 0.02–0.20 (Travis et al. 1996; Palacios and González-Candelas 1999; Palacios et al. 1999; Drummond et al. 2000; Gaudeul et al. 2000; Nuez et al. 2004).
Several life history and demographic factors, such as population size, breeding system, and phylogenetic position, are important in explaining genetic diversity within species and the apportionment of the diversity among populations of the species (Hamrick and Godt 1996; Weller et al. 1996; Hamrick and Godt 1997). Like other members of the woody Sonchus alliance, S. gandogeri is insect-pollinated and predominantly an outcrossing species (Aldridge 1975, 1979). Therefore, this breeding system may contribute to high genetic diversity compared to other endemics, especially Limonium dufourii and L. cavanillesii, which are apomictic species. Furthermore, S. gandogeri shows much higher genetic diversity than other insect-pollinated outcrossing species (e.g., Eryngium alpium, Astragalus cremnophylax var. cremnophylax). Both ITS and cpDNA phylogenies strongly suggest that S. gandogeri belongs to the lineage that radiated most recently within the woody Sonchus alliance (Kim et al. 1996b; S.-C. Kim, unpublished data). Also, El Hierro, where S. gandogeri occurs, is the youngest island (i.e., <1 Ma) in the Canaries. Therefore, molecular phylogenetic studies and the age of the island suggest that the old lineage hypothesis (Engler 1879; Bramwell 1976, 1990) is an unlikely explanation for the high genetic diversity of S. gandogeri. In addition, the population size of S. gandogeri is relatively small and the plants all occur on the almost inaccessible north-facing vertical cliffs. Despite small population sizes of S. gandogeri, the mean genetic diversity is much higher than other widely distributed endangered species (e.g., E. alpium).
It is possible that the long-lived woody habit may contribute to the high genetic diversity of the species. In general, long-lived woody perennials have higher genetic diversity at the population level than short-lived perennials and annuals (Hamrick et al. 1991; Hamrick and Godt 1997). At least within the woody Sonchus alliance (Kim et al. 1999), based on isozymes, there is a general trend of high genetic diversity in long-lived shrubs or trees (e.g., Babcockia, S. brachylobus, S. hierrensis, S. pinnatifidus; average HT=0.208) and low diversity in caudex perennials (e.g., S. acaulis, S. gummifer, S. gonzalezpadronii: average HT=0.049) or herbaceous perennials (e.g., S. tuberifer, Lactucosonchus: average HT=0.073). Therefore, it is plausible that life form contributes high genetic diversity to S. gandogeri despite its recent origin on El Hierro.
An alternative hypothesis to explain the high genetic diversity in the species is hybrid origin or introgressive hybridization. Hybridization in island groups generates genetic diversity in otherwise uniform island plants and could possibly lead to new species by stabilization of hybrid recombinants. High incidence of interspecific or even intergeneric hybridization has been documented in Canary Island endemics (Francisco-Ortega et al. 2000), and this may explain the high genetic diversity of S. gandogeri, as well as other endemics on the Canaries and Macaronesian islands. In contrast, interspecific hybridization is rare on the Juan Fernandez Islands and other Pacific archipelagos (Stuessy et al. 1998; Crawford et al. 2001). Aldridge (1976) hypothesized that S. gandogeri, well established on El Hierro, is probably a hybrid between S. pinnatus subsp. canariensis (=S. canariensis) and S. hierrensis. The chromosome number of S. gandogeri is 2n=18 (n=9), which is the same as other Dendrosonchus species in the Canaries (Rox and Boulos 1972). We need to further investigate a potential diploid hybrid speciation of this taxon based on both nuclear and chloroplast DNA. If indeed it is of hybrid origin, then it seems critical to determine whether geographically isolated populations have arisen once or multiple times.
Apportionment of diversity among populations
In addition to determining levels of diversity within species and populations, knowing how diversity is apportioned within and among populations of a species is useful in formulating strategies for conserving diversity within taxa (Hamrick et al. 1991). This study indicates that the majority of genetic diversity of Sonchus gandogeri populations is contained within populations, as indicated by the GST value of 0.149. Literature values obtained with AFLP and RAPD markers indicate that it is within the range of these markers: average RAPD GST value of 0.193 for 35 plant species (Bussel 1999) and AFLP range of 0.103–0.538 for endangered species (Travis et al. 1996; Palacios et al. 1999; Palacios and González-Candelas 1999; Gaudeul et al. 2000; Zawko et al. 2001; Nuez et al. 2004).
Absence of strong genetic differentiation can be explained by high gene flow (pollen-mediated or seed-mediated) or by the history of populations, such as recent colonization. The majority of Sonchus species on the islands have a strictly allogamous breeding system and appear to have an insect-mediated pollination mechanism: they are generally pollinated by small insects and/or butterflies (S.-C. Kim, personal observation). This may result in relatively frequent long-distance pollen dispersion, contributing to low genetic differentiation between the two populations. It is also possible that gene flow occurs through the movement of seeds. Achenes of S. gandogeri, like other taxa of Sonchus, have pappus with two different hair types, and this allows relatively easy dispersal of seeds on the island by strong winds, especially by predominant northeastern trade winds. Lastly, it is also plausible that the Las Esperillas population was recently colonized from the El Golfo population. Prevalent wind direction of the northeast trade winds, from north to south, in the Canaries suggests the possibility of this scenario. In addition, paraphyly of the El Golfo population indicated by NJ analysis (Fig. 2) and much lower genetic diversity of the Las Esperillas population support this hypothesis.
Conservation implications
Island populations and insular endemics are more prone to extinction than are mainland populations or species, due to genetic factors such as inbreeding depression, loss of genetic variation, and genetic adaptation to island conditions or to their interactions with demographic or environmental stochasticity, which are more severe than on the mainland (Ellstrand and Elam 1993; Frankham 1997, 1998). In addition, island populations are often small and few, and it is generally expected that small populations are more likely than larger ones to become extinct (Menges 1991).
Sonchus gandogeri seems less likely to become extinct due to genetic factors. Rather, environmental or demographic forces are more likely to lead to extinction. Although there are no direct threats from urban development and farming, the two populations are still threatened by introduced herbivores, such as goats. The goats can access vertical cliffs almost inaccessible by humans, and they have a detrimental impact on native flora. Therefore, more strict control of introduced herbivores is necessary to protect these populations.
For ex situ conservation, we need to carefully design and maintain a germplasm bank for this taxon. Seeds of Sonchus gandogeri, as well as other Sonchus taxa in the Canaries, can be kept refrigerated for several years, and germination rates remain high (S.-C. Kim, personal observation). These attributes are ideal for conservation of this taxon in seed banks. The El Golfo population, which has much higher genetic variability than the Las Esperillas one, must be given priority in the seed bank. However, this does not necessarily mean that we should neglect or not sample the Las Esperillas population. It is simply unknown what portion or proportion of genetic variation is responsible for adaptation to a changing environment. Given the limited distribution and the population size of this taxon, the collection of seeds from both the El Golfo and Las Esperillas populations is recommended in order to preserve all existing variability and for survival and reproduction of this species.
References
Aldridge AE (1975) Taxonomic and anatomical studies in Sonchus L. subg. Dendrosonchus Webb ex Schultz Bip. and related genera. PhD Thesis, University of Reading, Reading
Aldridge AE (1976) Macaronesian Sonchus subgenus Dendrosonchus s. l. (Compositae-Lactuceae), including a reappraisal of the species concept and new combinations. Bot Macaronesica 2:81–93
Aldridge AE (1979) Evolution within a single genus: Sonchus in Macaronesia. In: Bramwell D (ed) Plants and island. Academic Press, London, pp 249–285
Batista F, Bañares A, Caujapé-Castells J et al. (2001) Allozyme diversity in three endemic species of Cistus (Cistaceae) from the Canary Islands: intraspecific and interspecific comparisons and implications for genetic conservation. Am J Bot 88:1582–1592
Beardmore JA (1983) Extinction, survival, and genetic variation. In: Schoenwald-Cox CM, Chambers SM, MacBryde B, Thomas L (eds) Genetics and conservation. Benjamin-Cummings, Menlo Park, CA, pp 125–151
Boulos L (1974) Révision systematique du genre Sonchus L. s.l. V. Sous genre 2. Dendrosonchus. Botaniska Notiser 127:7–37
Bouza N, Caujapé-Castells J, González-Pérez MA, Batista F, Sosa PA (2002) Population structure and genetic diversity of two endangered endemic species of the Canarian laurel forest: Dorycnium spectabile (Fabaceae) and Isoplexis chalcantha (Scorphulariacaeae). Int J Plant Sci 163:619–630
Bramwell D (1976) The endemic flora of the Canary Islands: distrubution relationships and phytogeography. In: Kunkel G (ed) Biogeography and ecology in the Canary Islands, Monographiae Biologicae 30. Junk, The Hague, pp 207–240
Bramwell D (1990) Conserving biodiversity in the Canary Islands. Ann MO Botanical Garden 77:28–37
Bramwell D, Bramwell Z (2001) Wild flowers of the Canary Islands, 2nd edn. Editorial Rueda, Madrid
Bussel JD (1999) The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petrea (Lobeliaceae). Mol Ecol 8:775–789
Cardoso MA, Provan W, Powell J, Ferreras PCG, Oliveira DE (1998) High genetic differentiation among remnant populations of the endangered Caesalpinia echinata Lam. (Leguminosae-Caesalpinioideae). Mol Ecol 7:601–608
Carracedo JC (1994) The Canary Islands: an example of structural control on the growth of large oceanic-island volcanoes. J Volcanol Geotherm Res 60:225–241
Chalmers KJ, Waugh R, Sprent JI, Simons AJ, Powell W (1992) Detection of genetic variation between and within populations of Gliricidia sepium and G. maculata using RAPD markers. Heredity 69:465–472
Crawford DJ, Ruiz E, Stuessy TF et al. (2001) Allozyme diversity in endemic flowering plant species of the Juan Fernandez Archipelago, Chile: ecological and historical factors with implications for conservation. Am J Bot 88:2195–2203
De Joode DE, Wendel J (1992) Genetic diversity and origin of the Hawaiian Islands cotton, Gossypium tomentosum. Am J Bot 79:1311–1319
Drummond RSM, Keeling DJ, Richardson TE, Gardner RC, Wright SD (2000) Genetic analysis and conservation of 31 surviving individuals of a rare New Zealand tree, Metrosideros bartlettii (Myrtaceae). Mol Ecol 9:1149–1157
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Engler A (1879) Versuch einer Entwicklungsgeschichte, insbesondere der Florengebiete seit der Tertiärperiode. I. Die extratropischen Gebiete der nördlichen Hemisphäre. Engelmann, Leipzig
Francisco-Ortega A, Santos-Guerra A, Kim S-C, Crawford DJ (2000) Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87:909–919
Frankel OH (1983) The place of management in conservation. In: Schoenwald-Cox CM, Chambers SM, MacBryde B, Thomas L (eds) Genetics and conservation. Benjamin-Cummings, Menlo Park, CA, pp 125–151
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327
Frankham R (1998) Inbreeding and extinction: island populations. Conserv Biol 12:665–675
Franklin ER (1980) Evolutionary change in small populations. In: Soulé ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer, Sunderland, pp 135–150
Gaudeul M, Taberlet P, Till-Bottraud (2000) Genetic diversity in an endangered plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol Ecol 9:1625–1637
Gemmill CE, Ranker TA, Ragone D, Perlman SP, Wood KR (1998) Conservation genetics of the endangered endemic Hawaiian genus Brighmania (Campanulaceae). Am J Bot 85:528–539
Hamrick JL, Godt JW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics: case studies from nature. Chapman & Hall, NY, pp 281–301
Hamrick JL, Godt JW (1997) Effects of life history traits. In: Silvertown J, Franco M, Harper JL (eds) Plant life histories-ecology, phylogeny and evolution. Cambridge University Press, Cambridge, pp 102–118
Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991) Correlations between species traits and allozyme diversity: implications for conservation biology. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 75–86
Huenneke LF (1991) Ecological implications of genetic variation in plant populations. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 31–44
Kim S-C, Rieseberg LH (1999) Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics 153:965–977
Kim S-C, Crawford DJ, Jansen RK (1996a) Phylogenetic relationships among the genera of the subtribe Sonchinae (Asteraceae): evidence from ITS sequences. Syst Bot 21:417–432
Kim S-C, Crawford DJ, Francisco-Ortega J, Santos-Guerra A (1996b) A common origin for woody Sonchus and five related genera in the Macaronesian islands: molecular evidence for extensive radiation. Proc Natl Acad Sci USA 93:7743–7748
Kim S-C, Crawford DJ, Francisco-Ortega A, Santos-Guerra A (1999) Adaptive radiation and genetic differentiation in the woody Sonchus aliance (Asteraceaae: Sonchinae) in the Canary Islands. Plant Syst Evol 215:101–118
Manly BFJ (1985) The statistics of natural selection. Chapman & Hall, London
Menges ES (1991) The application of minimum viable population theory to plants. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 45–61
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nuez P, Prohens J, Blanca JM (2004) Relationships, origin, and diversity of Galápagos tomatoes: implications for the conservation of natural populations. Am J Bot 91:86–99
Palacios C, González-Candelas F (1999) AFLP analysis of the critically endangered Limonium cavanillesii (Plumbaginaceae). J Hered 90:485–489
Palacios C, Kresovich S, González-Candelas F (1999) A population genetic study of the endangered plant species Limonium dufourii (Plumbaginaceae) based on amplified fragment length polymorphism (AFLP). Mol Ecol 8:645–657
Rox J, Boulos L (1972) Révision systématique du genre Sonchus L. s. l. II. Étude caryologique. Botaniska Notiser 125:306–309
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schaal BA, Leverich WJ, Rogstad SH (1991) A comparison of methods for assessing genetic variation in plant conservation biology. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 123–134
Schmidt K, Jenssen K (2000) Genetic structure and AFLP variation of remnant populations in the rare plant Pedicularis palustris (Scrophulariaceae) and its relation to population size and reproduction components. Am J Bot 87:678–689
Stuessy TF, Crawford DJ, M Silva O (1998) Isolating mechanisms and modes of speciation in the vascular flora of the Juan Fernandez Islands. In: Stuessy TF, Ono M (eds) Evolution and speciation in island plants. Cambridge University Press, Cambridge, pp 79–86
Swofford DL (2000) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland
Travis SE, Maschinski J, Keim P (1996) An analysis of genetic variation in Astragalus cremnophylax var. cremnophylax, a critically endangered plant, using AFLP markers. Mol Ecol 5:735–745
Vos P, Hogers R, Bleeker M et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Weller SG, Sakai AK, Straub C (1996) Allozyme diversity and genetic identity in Schiedea and Alsinidendron (Caryophyllaceae: Alsinoideae) in the Hawaiian Islands. Evolution 50:23–34
Yeh FC, Young RC, Timothy B et al. (1997) Popgene, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Canada
Zawko G, Krauss SL, Dixon KW, Sivasithamparam K (2001) Conservation genetics of the rare and endangered Leucopogon obtectus (Ericaceae). Mol Ecol 10:2389–2396
Acknowledgements
The authors wish to thank Daniel Crawford, Javier Francisco-Ortega, and Norm Ellstrand for helpful comments on an early version of the manuscript. The manuscript was greatly improved by comments from two anonymous reviewers. This project is supported in part by an Academic Senate Grant, Regents’ Faculty Fellowship, and Agricultural Experiment Station funds from the University of California at Riverside to S.-C. Kim. We also thank Cabildo de El Hierro for issuing a permit to collect plants on El Hierro and the ICIA (Instituto Canario de Investigaciones Agrarias) for letting us use their facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, S.C., Lee, C. & Santos-Guerra, A. Genetic analysis and conservation of the endangered Canary Island woody sow-thistle, Sonchus gandogeri (Asteraceae). J Plant Res 118, 147–153 (2005). https://doi.org/10.1007/s10265-005-0203-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-005-0203-9