Abstract
Russian Far East loaches of the genera Cobitis and Misgurnus are among members of the family Cobitidae with poorly understood systematics. In this study we present phylogenetic hypotheses based on mitochondrial (cytochrome b) and nuclear (RAG-1) sequences. All analyses recovered comparable topological phylogenies, and all data sets supported the non-monophyly of the genera Cobitis and Misgurnus. Both genera are represented by multiple lineages that in some cases do not correspond to the species described. We found some phylogenetic incongruities for the genus Misgurnus (M. mohoity and M. anguillicaudatus) that are explained by ancient hybridization, as was suggested previously for M. anguillicaudatus. The revealed phylogenetic relationships suggest that Paramisgurnus should be treated as a synonym of Misgurnus and M. bipartitus as a synonym of M. mohoity. All analyses recovered C. choii as a member of the genus Cobitis, confirming previous taxonomic conclusions. Most of the molecular lineages found follow currently recognized taxa with some exceptions, such as M. anguillicaudatus and C. lutheri. Phylogenetic relationships recover several unrelated lineages of M. anguillicaudatus and suggest additional studies to solve current taxonomic uncertainty. We found that C. lutheri is a non-natural group that contains two unrelated lineages: specimens of C. lutheri from the Far East of Russia collected close to the type locality and a second lineage with specimens of C. lutheri from Korea, the identification of which must be revised. The study provides evidence of the presence of the Far East species M. nikolskyi in Sakhalin Island, but simultaneously shows conspicuous genetic distinctiveness between the island and the mainland populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Far East Region of Russia represents the northeastern extreme of Asia with river drainages emptying towards the Pacific Ocean. It is located east of the Siberian and Baikal Lake areas and includes Sakhalin, Kuril, Wrangel, the Commander, and Shantarskiye islands. This region represents an area of faunal transition without major faunal breaks between Asian (southern) and Palearctic (northern) realms with a specific taxonomic composition of freshwater fish fauna (Warpachowski and Herzenstein 1887; Berg 1909; Taranetz 1938; Chereshnev 1998; Bogutskaya et al. 2008). The Amur River with more than 200 tributaries is the main river system of the Far East region of Russia, covering also territories of northern China and northeastern Mongolia. Although recent studies have solved taxonomic problems in several taxa (Vasil’eva and Kozlova 1989; Vasil’eva 2001, 2007; Stevenson 2002; Shedko and Shedko 2003; Vasil’eva and Makeyeva 2003; Vasil’eva et al. 2003; Shedko and Chereshnev 2005; Shedko et al. 2005, 2008; Knizhin et al. 2006, 2007; Yamazaki et al. 2006; Vasil’ev and Vasil’eva 2008a, b), there are several groups of freshwater fishes with uncertain taxonomy and distribution limits in the Far East of Russia.
Recent studies on loaches of the genera Cobitis and Misgurnus have shown that three species of the genus Cobitis, namely Cobitis choii, Cobitis lutheri, and Cobitis melanoleuca, and two Misgurnus species, Misgurnus mohoity and Misgurnus nikolskyi, occur in the Amur River drainage and coastal waters of the Far East region of Russia (Vasil’eva 1998, 2001; Vasil’eva et al. 2003; Vasil’ev and Vasil’eva 2008a, b). Most of these loach species are restricted to East Asia, while C. melanoleuca is the only spined loach with continuous distribution from the Russian Far East and China to Europe with the western populations in the Don and Kuban river basins (Vasil’eva 1998). In the Far East region of Russia, C. melanoleuca is represented by the nominotypical subspecies C. m. melanoleuca that differs in the karyotype structure from the Siberian (C. m. granoei) and European (C. m. gladkovi) subspecies (Vasil’ev and Vasil’eva 2008a). Despite great progress in the taxonomy of loaches from the Far East of Russia, several systematic issues at the specific and population levels remain unsolved and need to be clarified. In recent years, molecular studies have elucidated the phylogenetic relationships of loaches of the family Cobitidae (Tang et al. 2006; Šlechtová et al. 2008). Their major conclusions showed many genera of the Cobitidae as non-monophyletic groups, e.g., Cobitis and Misgurnus, and in many taxa there was significant disparity between morphological and molecular results. These conclusions reinforce the idea that early classifications that relied on morphological characters, e.g., barbels, scale of Canestrini, scale, and pigmentation, to define taxonomic boundaries were not phylogenetic and in many cases were based on subjective identification of taxonomic differentiation. The other major conclusion was the identification of introgressed mtDNA in different loach groups, such as the genus Misgurnus, based on the incongruence of molecular markers (Šlechtová et al. 2008).
In this study, we used mitochondrial (cytochrome b) and nuclear (RAG-1) genes of all Russian Far East loaches of the genera Cobitis and Misgurnus, and other related genera to infer their phylogenetic relationships. We used range-wide populations of the different species of Cobitis and Misgurnus from the Far East of Russia and adjacent areas to evaluate their genetic variability and to identify their intraspecific structure. We analysed several morphologically atypical populations to provide a wide framework for evaluating contentious loach taxa, and we suggested an alternative hypothesis for the systematics of the genus Misgurnus.
Materials and methods
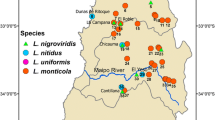
Specimens analysed. All loach species were collected by hand nets in 24 localities of the Far East region of Russia from 1996 up to 2007 (Fig. 1; Table 1). Thirty-one new specimens of Cobitis and 21 new Misgurnus were sequenced for cytochrome b (cyt b) and RAG-1. We included groups phylogenetically related with the Far East loaches (Šlechtová et al. 2008): Paramisgurnus, Koreocobitis, Niwaella, and Sabanejewia plus a wide representation of Misgurnus—Misgurnus anguillicaudatus, Misgurnus bipartitus, Misgurnus fossilis, and Misgurnus mizolepis, as well as Misgurnus sp. 1, Misgurnus sp. 2, and Misgurnus sp. 3 (as defined in the study of Šlechtová et al. 2008), and Misgurnus sp. (as in Wang and Tzeng, unpublished data), and Cobitis—Cobitis biwae, Cobitis hankugensis, Cobitis rara, Cobitis shikokuenis, Cobitis sinensis, Cobitis striata, Cobitis takatsuensis, and Cobitis misgurnoides. Currently C. misgurnoides is not considered a member of the genus Cobitis, and it was erected as the new genus Microcobitis by Bohlen and Harant (2010). Microcobitis was used as a close outgroup, whereas Pangio and Sabanejewia were phylogenetically more distant outgroups (Šlechtová et al. 2008). These sequences were recovered from GenBank [see Electronic Supplementary Material (ESM) Table S1] and new sequences were deposited in GenBank. Accession numbers (JN858807–858899) and a list of specimens sequenced with localities and collection numbers are provided in Table 1.
Map indicating locations of sampling sites (numbered 1–24) for studied Misgurnus and Cobitis species from the Far East of Russia. Letters designate species according to the codes from Table 1
PCR analysis. Total DNA was extracted from ethanol-preserved fin tissue using Charge Switch gDNA Microtissue Kit (Invitrogen Inc., Carlsbad, CA, USA). The entire cyt b (1,140 base pair; bp) was PCR amplified using the primers GluDGL (Palumbi 1996) and H16460, or the primers Glu-L.Ca14337–14359 and Thr.-H.Ca15568–15548 (Šlechtová et al. 2006). Nuclear RAG-1 (897 bp) was PCR amplified using the primers RAG1-1F (Quenouille et al. 2004) and RAGRV1 (Šlechtová et al. 2007). Both genes were amplified in 25-μl PCR reactions using the conditions described in Perdices et al. (2008). Primers for sequencing the purified PCR were the same as those used for PCR amplifications. Chromatograms and alignments were revised using Sequencher ver. 4.8 (Gene Codes Corporation Inc., Ann Arbor, MI, USA). The final data set for cyt b contained 1,114 bp to make comparable sequence alignments with some published sequences. None of the genes exhibited stop codons or gaps for cyt b or RAG-1, and all positions were used in the phylogenetic analyses. Nucleotide composition and base frequencies for all positions were checked using PAUP* 4.0b10 (Swofford 2002). Sequence divergences were calculated using Sequencer 6.1 (written by B. Kessing).
Phylogenetic analysis. Phylogenetic analyses were conducted for each aligned data set using the Bayesian inference (BI) method as implemented in MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001), neighbour joining (NJ), and maximum parsimony (MP) methods using PAUP*. MP analysis was performed by heuristic searches with TBR branch swapping and ten replicates of random addition of taxa. Only minimal trees were retained and the zero branch length collapsed. For the NJ method, we selected the best fit nucleotide substitution model for each individual data set using the Akaike information criterion (AIC) by Modeltest ver. 3.7 (Posada and Crandall 1998). For the BI method, we ran 3,000,000 cycles of four simultaneous Monte Carlo Markov chains, sampling the Markov chains at intervals of 100 generations. Log-likelihood stability was attained after 80,000 generations, and we excluded the first 800 trees as burn-in. The remaining trees were used to compute a 50% majority rule consensus tree. Robustness of the inferred trees in the MP and NJ analyses was assessed by bootstrapping (1,000 replicates) (Felsenstein 1985), and by posterior probability values (ppb) in the BI procedure.
Results
Sequence diversity. The cytochrome b (1,114 bp) and nuclear RAG-1 (897 bp) nucleotide sequences were obtained from 52 new individuals (Table 1). All cyt b and RAG-1 sequences showed similar base composition each. Cytochrome b sequences had a strong bias against guanine (15.0%), a situation not observed in the nuclear gene (27.5%). Plots of transitions and transversions against uncorrected p-distances showed an absence of nucleotide saturation for cyt b and RAG-1. Of the 1,114 bp for cyt b, 491 were variable (11.7% 1st position, 2.5% 2nd, 32.4% 3rd) and 452 parsimony informative; for RAG-1, 178 positions were variable (19.8%) and 135 parsimony informative (15.1%) (excluding outgroups). Mean uncorrected p-distances found among congeneric species were 14.0 ± 2.3% for cyt b and 2.5 ± 0.5% for RAG-1 for Cobitis, and 14.7 ± 3.0% for cyt b and 2.1 ± 0.8% for RAG-1 for Misgurnus. Modeltest identified the GTR + G (1.1276) + I (0.5085) model as the most appropriate model for cyt b, and the SYM + G (0.8143) + I (0.6280) model for the RAG-1.
Phylogenetic relationships. All analyses (BI, MP, and NJ) were highly congruent for the separate cyt b and RAG-1 data sets. Although we found some incongruities between mitochondrial and nuclear topologies, both data sets supported the non-monophyly of the genera Cobitis and Misgurnus (Figs. 2, 3). Both Cobitis and Misgurnus are represented by multiple lineages that in some cases do not correspond to species described for both genera.
Phylogenetic relationships based on cyt b sequences. Bayesian tree (50% majority rule consensus) using the GTR + I + G model, with values on branches corresponding to Bayesian posterior probabilities, and NJ and MP bootstrap values. An asterisk appears when all values were 100%, and a dash appears when the branch is not supported. Dotted branches identified specimens of Misgurnus anguillicaudatus, and grey branches identified specimens of Niwaella
Phylogenetic relationships based on RAG-1 sequences. Bayesian tree (50% majority rule consensus) using SYM + I + G model, with values on branches corresponding to Bayesian posterior probabilities, and NJ and MP bootstrap values. An asterisk appears when all values were 100%, and a dash appears when the branch is not supported. Dotted branches identified specimens of Misgurnus anguillicaudatus, and grey branches identified specimens of Niwaella
Mitochondrial analyses showed two major clades well supported in BI (98% ppb) (Fig. 2). One major clade, Clade A (100% ppb, >66% bootstrap MP, NJ), included all Cobitis species, Niwaella delicata, N. multifasciata, and some species of Misgurnus (M. anguillicaudatus, M. bipartitus, M. mohoity, Misgurnus sp., Misgurnus sp. 2, Misgurnus sp. 3) (Fig. 2). Samples of Niwaella were related with different Cobitis species. The specimens of C. lutheri from the Far East of Russia and Korea were not recovered as a monophyletic group. They were separated in two independent lineages with geographical structure and high genetic distances (mean uncorrected p-distances 11.3 ± 0.25%). All specimens of C. lutheri from the Far East of Russia and one specimen from China were recovered in one linage, while all specimens from Korea formed a different lineage closely related to C. striata (Biwa small race). Other Russian Far East loaches, C. melanoleuca and C. choii, were always recovered as monophyletic with low genetic intraspecific divergences (0.2 ± 0.2 and 0.2 ± 0.2%, respectively).
The species C. biwae, C. striata, and M. anguillicaudatus were represented by multiple lineages, not related in a monophyletic group. These three taxa are documented as hybrid species with different ploidy (Kitagawa et al. 2003; Morishima et al. 2008; Šlechtová et al. 2008; Saitoh et al. 2010). Their phylogenetic relationships, as already shown in previous studies, varied on their ploidy and on the mitochondrial or nuclear gene used (see Saitoh et al. 2000, 2010; Kitagawa et al. 2001, 2003, 2005). Specimens of M. mohoity, M. bipartitus, and Misgurnus sp. 3 were recovered in a monophyletic group, and closely related to some specimens of M. anguillicaudatus and Misgurnus sp. 2.
The second major clade, Clade B, supported in BI included together Koreocobitis, Paramisgurnus, and the rest of the Misgurnus species analysed. Samples of M. mizolepis were recovered among specimens of P. dabryanus with relatively low genetic distances (uncorrected p-distances 2.5 ± 1.3%). We found high intraspecific genetic diversity in M. nikolskyi specimens (p-distances 6.3 ± 4.4%) with at least two well-differentiated mitochondrial lineages. One of them included all individuals collected on Sakhalin Island, and some specimens identified as M. anguillicaudatus from Japan, and another lineage that related M. nikolskyi specimens collected in the Far East of Russia, Misgurnus sp. 1 (Šlechtová et al. 2008), and one specimen from the Amur River in China (Saitoh et al. 2006). The species M. fossilis was recovered as monophyletic with low genetic divergences among specimens (0.2 ± 0.3%). In MP and NJ analyses, the second major clade, Clade B, was not supported, and it was subdivided into three independent lineages: one lineage included Paramisgurnus and Misgurnus mizolepis, the second lineage corresponded to Koreocobitis, and the third lineage included the rest of the Misgurnus species.
In general, nuclear phylogenies are less resolved than mitochondrial topologies (Fig. 3). We found two major clades with a moderate-high bootstrap value (>56%) and ppb (98%), which do not correspond exactly with the mitochondrial phylogeny. Clade A included exclusively Cobitis species and Niwaella delicata and N. multifasciata. Similarly to the mitochondrial results, C. lutheri was not monophyletic. All C. lutheri individuals from the Far East of Russia comprise a well-supported monophyletic clade distanct from C. lutheri specimens from Korea (2.1 ± 0.2%), and C. lutheri specimens from Korea were related to C. striata (Large and Middle races). The specimens of Niwaella species were related to different Cobitis species as in the mitochondrial phylogeny. Clade B included all species of Misgurnus analysed, Koreocobitis and Paramisgurnus. Koreocobitis was always basal, and M. mizolepis was always embedded within Paramisgurnus as in the mitochondrial phylogeny (mean p-distances 0.2 ± 0.1%). Misgurnus mohoity was recovered as monophyletic, and it was related to Misgurnus sp. 2 from Korea, and to some samples identified as M. anguillicaudatus from Korea and Japan and Misgurnus sp. 3 from Korea, as in the mitochondrial topology. Misgurnus fossilis was recovered as non-monophyletic in MP and NJ analyses, and with low support in Bayesian analysis for RAG-1 (Fig. 3).
Discussion
Genera Cobitis and Misgurnus and related groups. Mitochondrial and nuclear analyses of the Russian Far East loaches and close relatives produced, at some levels, incongruent phylogenetic results. In all analyses, the genera Misgurnus and Cobitis were paraphyletic. These results indicate that morphological variation used for previous taxonomic hypotheses does not reflect phylogenetic relationships among members of these genera. Relationships described by cyt b and RAG-1 phylogenies always produced non-monophyletic groups for the genera Cobitis and Misgurnus. In our analyses, some species of Niwaella and Paramisgurnus were always intimately related to Cobitis and Misgurnus, respectively. Niwaella delicata was always phylogenetically more closely related to Cobitis species than to other Niwaella species; N. multifasciata. Misgurnus mizolepis was embedded in the Paramisgurnus dabryanus lineage. Therefore, neither Cobitis nor Misgurnus are natural groups as currently recognised, as has been suggested in other studies (Tang et al. 2006; Šlechtová et al. 2008).
Mitochondrial and nuclear incongruities were especially relevant in the relationships of some Misgurnus species with the Cobitis group. The close mitochondrial relation found among M. mohoity and M. anguillicaudatus and some Cobitis species suggests that Russian Far East M. mohoity is also introgressed at the mitochondrial level, as was previously suggested for M. anguillicaudatus (Šlechtová et al. 2008). On the basis of the strong differences of the mitochondrial and nuclear relationships observed, Šlechtová et al. (2008) suggested a past hybridization of M. anguillicaudatus and a member of Cobitis with subsequent backcrosses of the hybrid with the parental species. Our results also indicated strong phylogenetic differences at the mitochondrial and nuclear levels for M. mohoity and M. anguillicaudatus, and we suggest the hybridization as a possible mechanism for explaining these incongruities.
At the nuclear level, all Misgurnus and Paramisgurnus species were grouped in a monophyletic clade that related all Asiatic species with the Central European M. fossilis as the basal member of all Misgurnus and Paramisgurnus. In all nuclear topologies Koreocobitis was the sister group to this clade. The recovery of M. mizolepis embedded within the P. dabryanus lineage is consistent with the argument that M. mizolepis represents a junior synonym of P. dabryanus (Vasil’eva 2001).
Concerning the systematics of the genus Misgurnus, two alternative hypotheses could be proposed. (1) The relationships of P. dabryanus with other Misgurnus species suggest that Paramisgurnus should be considered as a member of the genus Misgurnus (Šlechtová et al. 2008). The inclusion of P. dabryanus in the genus Misgurnus will convert this genus in a monophyletic group, as was shown in the molecular phylogenies (Fig. 3). (2) According to our results, the acceptance of the monotypic genus Paramisgurnus implies the restriction of the genus Misgurnus to its type species, M. fossilis, and therefore, Asian Misgurnus must be considered a new genus. Misgurnus fossilis is the only non-Asiatic member of the genus Misgurnus, and it was suggested that this species was a Pliocene immigrant from East Asia (Bănărescu 1990). Our molecular phylogenies indicate that the M. fossilis lineage is deeply divergent from Asian Misgurnus and Paramisgurnus, and therefore it is difficult to concur with the hypothesis of a Pliocene immigrant. Further work based on multiple types of evidence (genetics, morphology, karyology) must resolve current systematic delimitation of the genus and species of Misgurnus.
Systematic implication. The molecular characterization of the Russian Far East species of the genus Misgurnus showed that all specimens of M. mohoity from the middle and lower Amur drainage had low intraspecific molecular divergence. At the mitochondrial level M. mohoity showed low genetic divergence from mud loaches identified as M. bipartitus by Tang et al. (2006), and some mud loaches from China and Korea identified as M. anguillicaudatus (Fig. 2; ESM Table S1). Earlier karyological and morphological studies have considered M. bipartitus a synonym of M. mohoity (Vasil’eva 2001; Vasil’eva et al. 2003). Our results confirm this conclusion and permit extending the range of M. mohoity to Korea from its known distribution in the Russian part of the Amur River drainage (except Khanka Lake), northeastern Mongolia, and northeastern China (south to the upper stream of the Liao River) (Vasil’eva et al. 2003). Although some M. anguillicaudatus from China should be treated as M. mohoity, M. anguillicaudatus continues to be paraphyletic with different well-defined lineages that probably correspond to different species. It has already been suggested that M. anguillicaudatus represents more than a single species with several phylogenetic lineages grouped under the same name (Khan and Arai 2000; Tang et al. 2006; Šlechtová et al. 2008; Vasil’ev and Vasil’eva 2008a, b). All recent evidence warrants the systematic revision of mud loaches currently considered as M. anguillicaudatus as well as other members of the genus Misgurnus.
The recovery of M. nikolskyi specimens from the mainland and those from Sakhalin Island as a distinct evolutionary lineage, respectively (see Table 1; Fig. 1) supports previous studies that have found some karyological and ecological variability for this species (Vasil’eva 2001; Vasil’eva et al. 2003). However, we found more genetic differentiation within this clade than expected. At the mitochondrial level, we found a core group of M. nikolskyi specimens from nearly all Far East localities of Russia and Misgurnus sp. 1 from Russia. This lineage is closely related to some individuals of M. nikolskyi recovered from GenBank from the Amur River in China. We recovered another well-differentiated lineage of M. nikolskyi with individuals from Sakhalin Island, mitochondrially related to some Japanese individuals identified as M. anguillicaudatus. At the nuclear level, the M. nikolskyi specimens from Sakhalin Island were not related in a monophyletic group with the continental specimens.
Therefore, the considerable genetic divergence of the Sakhalin lineage and its relationship with some Japanese mud loach individuals must be verified by more representative phylogeographic analyses. Morphological characters supported the identification of M. nikolskyi individuals analysed; however, a taxonomic study of Japanese mud loaches M. anguillicaudatus and M. nikolskyi is needed, as several of the specimens of both species are intimately related. This conclusion was also supported in previous studies that showed the genetic heterogeneity of Japanese mud loaches (Morishima et al. 2008; Koizumi et al. 2009).
The molecular phylogenies support non-monophyly of the genera Cobitis and Niwaella, as previously suggested by Šlechtová et al. (2008). All phylogenies show the species of Niwaella closely related to different species of Cobitis. The nested position of C. choii within all other species of Cobitis, as previously revealed by Šlechtová et al. (2008), provides support for considering this species as a member of the genus Cobitis (Vasil’ev and Vasil’eva 2008a, b; Kim 2009), and not a member of the genus Iksookimia (Kim et al. 1999; Kim and Park 2002; Kottelat 2006).
The molecular analysis of the Far East Cobitis shows C. lutheri from different localities as a non-natural group. Our results support two well-differentiated molecular lineages. One lineage related all C. lutheri specimens from the Far East of Russia with low genetic variability. The second lineage was recovered with Korean specimens identified as C. lutheri by Šlechtová et al. (2008) and Lee (2009), but phylogenetically related to C. striata from Japan (Figs. 2, 3). Our results support previous karyological results about the non-conspecificity of the Korean spined loaches identified as C. lutheri and C. lutheri s. stricto described from Khanka Lake (Vasil’ev and Vasil’eva 2008a, b). Therefore, we maintain C. lutheri s. stricto for the Russian Far East specimens that according to previous karyological studies also inhabit the rest of the Amur River basin and waters of Primorye district neighboring the type locality represented in our study by the sample from the Khanka Lake basin (locality 18 in Fig. 1). The close phylogenetic relationship of C. lutheri from Korea with another Korean species, C. tetralineata (Kitagawa et al. 2005), and the similar karyotype of the Korean C. lutheri with C. tetralineata, might suggest their consideration of local populations of this last species (Vasil’ev and Vasil’eva 2008a, b). Therefore, previous knowledge coupled with our phylogenetic results suggests that Korean specimens should be taxonomically revised.
The obtained molecular phylogenies do not support the taxonomic treatment of C. melanoleuca as a complex of three subspecies with karyological differences (Vasil’ev and Vasil’eva 2008a). Our results are more consistent with explanations that show C. melanoleuca as rather homogeneous species with very low genetic divergence across a broad geographic range. However, our study includes a restricted number of localities and should be considered as a preliminary result. Undoubtedly, more C. melanoleuca specimens need to be studied to support or not the presumed taxonomic distinctiveness of C. melanoleuca subspecies.
References
Bănărescu P (1990) Zoogeography of fresh waters. Vol. 1. General distribution and dispersal of freshwater animals. AULA-Verlag GmbH, Wiesbaden
Berg LS (1909) Fishes of the Amur River. Zap Imp Akad Nauk Fiz-Mat Otd 24:1–270
Bogutskaya NG, Naseka AM, Shedko SV, Vasil’eva ED, Chereshnev IA (2008) The fishes of the Amur River: updated check-list and zoogeography. Ichthyol Explor Freshw 19:301–366
Bohlen J, Harant R (2010) Microcobitis, a new genus name for Cobitis misgurnoides (Teleostei: Cobitidae). Ichthyol Explor Freshw 21:295–300
Chereshnev IA (1998) Biogeography of freshwater fishes of the Far East in Russia. Dal’nauka, Vladivostok
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Khan MR, Arai K (2000) Allozyme variation and genetic differentiation in the loach Misgurnus anguillicaudatus. Fish Sci 66:211–222
Kim IS (2009) A review of the spined loach, family Cobitidae (Cypriniformes) in Korea. Korean J Ichthyol Suppl 21:7–28
Kim IS, Park JY (2002) Freshwater fishes of Korea. Kyo-Hak Publishing Co, Seoul
Kim IS, Park JY, Nalbant T (1999) The Far-East species of the genus Cobitis with the description of three new taxa (Pisces: Ostariophysi: Cobitidae). Trav Mus d’Histoire Nat “Grigore Antipa” 41:373–391
Kitagawa T, Watanabe M, Kobayashi T, Yoshioka M, Kashiwagi M, Okazaki T (2001) Two genetically divergent groups in the Japanese spined loach, Cobitis takatsuensis, and their phylogenetic relationships among Japanese Cobitis inferred from mitochondrial DNA analyses. Zool Sci 18:249–259
Kitagawa T, Watanabe M, Kitagawa ET, Yoshioka M, Kashiwagi M, Okazaki T (2003) Phylogeography and the maternal origin of the tetraploid form of the Japanese spined loach, Cobitis biwae, revealed by mitochondrial DNA analysis. Ichthyol Res 50:318–325
Kitagawa T, Jeon SR, Kitagawa E, Yoshioka M, Kashiwagi M, Okazaki T (2005) Genetic relationships among the Japanese and Korean striated spined loach complex (Cobitidae: Cobitis) and their phylogenetic position. Ichthyol Res 52:111–122
Knizhin IB, Antonov AL, Weiss SJ (2006) A new subspecies of the Amur grayling Thymallus grubii flavomaculatus ssp. nova (Thymallidae). J Ichthyol 46:555–562
Knizhin IB, Antonov AL, Safronov SN, Weiss SJ (2007) New species of grayling Thymallus tugarinae sp. nova (Thymallidae) from the Amur River basin. J Ichthyol 47:123–139
Koizumi N, Takemura T, Watabe K, Mori A (2009) Genetic variation and diversity of Japanese loach inferred from mitochondrial DNA—phylogenetic analysis using the cytochrome b gene sequences. Trans JSIDRE 259:7–16
Kottelat M (2006) Fishes of Mongolia. A check list of the fishes known to occur in Mongolia with comments on systematics and nomenclature. The World Bank, Washington
Lee IR (2009) Studies on the conservation biology of an endangered freshwater fish, Iksookimia choii. Ph.D. Thesis, Soonchunhyang University, Asan
Morishima K, Nakamura-Shiokawa Y, Bando E, Li YJ, Boron A, Khan MMR, Arai K (2008) Cryptic clonal lineage and genetic diversity in the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) inferred from nuclear and mitochondrial analyses. Genetica 132:159–171
Palumbi S (1996) Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn. Sinauer, Sunderland, MA
Perdices A, Bohlen J, Doadrio I (2008) The molecular diversity of Adriatic spined loaches (Teleostei, Cobitidae). Mol Phylogenet Evol 46:382–390
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Quenouille B, Bermingham E, Planes S (2004) Molecular systematics of the damselfishes (Teleostei: Pomacentridae): Bayesian phylogenetic analyses of mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 31:66–88
Saitoh K, Kobayashi T, Ueshima R, Numachi KI (2000) Analyses of mitochondrial and satellite DNAs on spined loaches of the genus Cobitis from Japan have revealed relationships among populations of three diploid–tetraploid complexes. Folia Zool 49 (Suppl 1):3–7
Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M (2006) Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol 63:826–841
Saitoh K, Chen WJ, Mayden RL (2010) Extensive hybridization and tetrapolyploidy in spined loach fish. Mol Phylogenet Evol 56:1001–1010
Shedko SV, Chereshnev IA (2005) Review of the gobies (Perciformes, Gobiidae) from fresh waters of the Kuril Islands. In: Makarchenko EA (ed) Vladimir Ya. Levanidov’s Biennial Memorial Meetings 3. Dal’nauka, Vladivostok, pp 435–455
Shedko SV, Shedko MB (2003) New data on freshwater fish fauna of the south of the Far East of Russia. In: Makarchenko YeA, Bogatov VV, Tiunova TM, Medvedeva LA, Semenchenko AYu (eds) Vladimir Ya. Levanidov’s Biennial Memorial Meetings 2. Dal’nauka, Vladivostok, pp 319–336
Shedko SV, Shedko MB, Pietsch W (2005) Pungitius polyakovi sp. n., a new species of ninespine stickleback (Gasterosteiformes, Gasterosteidae) from south-eastern Sakhalin Island. In: Storozhenko Yu (ed) Flora and fauna of Sakhalin Island Pt 2. Dal’nauka, Vladivostok, pp 223–233
Shedko SV, Miroshnichenko IL, Nemkova GA (2008) On the systematics and phylogeography of eight-barbel loaches of the genus Lefua (Cobitoidea: Nemacheilidae): mtDNA typing of L. pleskei. Russ J Genet 44:817–825
Šlechtová V, Bohlen J, Freyhof J, Ráb P (2006) Molecular phylogeny of the Southeast Asian freshwater fish family Botiidae (Teleostei: Cobitoidea) and the origin of polyploidy in their evolution. Mol Phylogenet Evol 39:529–541
Šlechtová V, Bohlen J, Tan HH (2007) Families of Cobitoidea (Teleostei; Cypriniformes) as revealed from nuclear genetic data and the position of the mysterious genera Barbucca, Psilorhynchus, Serpenticobitis and Vaillantella. Mol Phylogenet Evol 44:1358–1365
Šlechtová V, Bohlen J, Perdices A (2008) Molecular phylogeny of the freshwater fish family Cobitidae (Cypriniformes: Teleostei): Delimitation of genera, mitochondrial introgression and evolution of sexual dimorphism. Mol Phylogenet Evol 47:812–831
Stevenson DE (2002) Systematics and distribution of fishes of the Asian goby genera Chaenogobius (Osteichthyes: Perciformes: Gobiidae), with the description of a new species. Spec Diversity 7:251–312
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer, Sunderland, MA
Tang Q, Liu H, Mayden R, Xiong B (2006) Comparison of evolutionary rates in the mitochondrial DNA cytochrome b gene and control region and their implications for phylogeny of the Cobitoidea (Teleostei: Cypriniformes). Mol Phylogenet Evol 39:347–357
Taranetz AYa (1938) On zoogeography of the Amur Transitional Province on the basis of freshwater fish fauna studies. Vestnik DV filiala Akadem Nauk SSSR 32(5):99–115
Vasil’ev VP, Vasil’eva ED (2008a) Comparative karyology of species of the genera Misgurnus and Cobitis (Cobitidae) from the Amur River Basin in connection with their taxonomic relations and the evolution of karyotypes. J Ichthyol 48:1–13
Vasil’ev VP, Vasil’eva ED (2008b) Comparative karyological analysis of mud loach and spined loach species (genera Misgurnus and Cobitis) from the Far East region of Russia. Folia Zool 57:51–59
Vasil’eva ED (1998) Fam. 16. Cobitidae. In: Reshetnikov YuS (ed) Annotated check-list of Cyclostomata and fishes of the continental waters of Russia. Nauka, Moscow, pp 97–103
Vasil’eva ED (2001) Loaches (genus Misgurnus, Cobitidae) of Russian Asia. I. The species composition in waters of Russia (with a description of a new species) and some nomenclature and taxonomic problems of related forms from adjacent countries. J Ichthyol 41:553–563
Vasil’eva ED (2007) Gobies of the genus Rhinogobius (Gobiidae) from Primor’e and water bodies of Central Asia and Kazakhstan: I. Morphological characteristic and taxonomic status. J Ichthyol 47:691–700
Vasil’eva ED, Makeyeva AP (2003) Taxonomic status of the black Amur bream and some remarks on problems of taxonomy of the genera Megalobrama and Sinibrama (Cyprinidae, Cultrinae). J Ichthyol 43:582–597
Vasil’eva ED, Vasil’ev VP, Skomorokhov MO (2003) Loaches (Misgurnus, Cobitidae) of Russian Asia: II. Morphology, synonymy, diagnoses, karyology, biology, and distribution. J Ichthyol 7:501–510
Vasil’eva ED, Kozlova MS (1989) Taxonomy of the sawbelly genus Hemiculter in the Soviet Union. J Ichthyol 29:123–135
Warpachowski NA, Herzenstein SM (1887) Notes on ichthyology of the Amur River basin and adjacent countries. Trudy Sanct-Petersburg obshchestva estestvoispytat Otd zoolog fisiolog 19:1–58
Yamazaki Y, Yokoyama R, Nishida M, Goto A (2006) Taxonomy and molecular phylogeny of Lethenteron lampreys in eastern Eurasia. J Fish Biol 68:251–26
Acknowledgments
This study was partially supported by project ref. no. CGL2007-61010 of the Ministerio de Ciencia e Innovación (Spain), by the Russian Foundation for Basic Research (grant no. 09-04-00211), by the Programs of the Presidium of the Russian Academy of Sciences “Dynamics of Gene Pools of Populations”, and the Program of Complex Investigations in the Amur River Basin FEB RAS. We thank P. Ochoa for the molecular work done at the Laboratory of Molecular Systematics at the Museo Nacional de Ciencias Naturales, CSIC (Madrid (Spain), and Dr. S.N. Safronov from Sakhalin University for the specimens of mud loaches from Sakhalin Island. We are grateful to Dr. K. Saitoh and two anonymous reviewers for comments that improved this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Perdices, A., Vasil’ev, V. & Vasil’eva, E. Molecular phylogeny and intraspecific structure of loaches (genera Cobitis and Misgurnus) from the Far East region of Russia and some conclusions on their systematics. Ichthyol Res 59, 113–123 (2012). https://doi.org/10.1007/s10228-011-0259-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10228-011-0259-6