Abstract
Objectives
Whole genome sequencing (WGS) is an emerging tool in clinical diagnostics. However, little has been said about its procedure costs, owing to a dearth of related cost studies. This study helps fill this research gap by analyzing the execution costs of WGS within the setting of German clinical practice.
Methodology
First, to estimate costs, a sequencing process related to clinical practice was undertaken. Once relevant resources were identified, a quantification and monetary evaluation was conducted using data and information from expert interviews with clinical geneticists, and personnel at private enterprises and hospitals. This study focuses on identifying the costs associated with the standard sequencing process, and the procedure costs for a single WGS were analyzed on the basis of two sequencing platforms—namely, HiSeq 2500 and HiSeq Xten, both by Illumina, Inc. In addition, sensitivity analyses were performed to assess the influence of various uses of sequencing platforms and various coverage values on a fixed-cost degression.
Results
In the base case scenario—which features 80 % utilization and 30-times coverage—the cost of a single WGS analysis with the HiSeq 2500 was estimated at €3858.06. The cost of sequencing materials was estimated at €2848.08; related personnel costs of €396.94 and acquisition/maintenance costs (€607.39) were also found. In comparison, the cost of sequencing that uses the latest technology (i.e., HiSeq Xten) was approximately 63 % cheaper, at €1411.20.
Conclusions
The estimated costs of WGS currently exceed the prediction of a ‘US$1000 per genome’, by more than a factor of 3.8. In particular, the material costs in themselves exceed this predicted cost.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Whole genome sequencing (WGS) is an emerging diagnostic tool, and it has the potential to generate an incomparable variety of genetic information. Individual genomes and genetic variations within the population can be characterized by genetic analyses [1]. In recent years, a better understanding of the relationship between genotype and phenotype has been achieved by conducting genome-wide association studies [2]. Hence, this genetic research, in concert with current technological progress, has provided the prerequisites for a broad application of genetic diagnostics (e.g., WGS) in medical care.
On account of continuous technological progress, significant cost reductions with respect to DNA sequencing have been realized over time [3]. This cost degression has been facilitated by the transition from the classic chain termination method (‘Sanger method’ [4]) to next-generation sequencing (NGS) technologies [1, 5]. The massively parallel sequencing inherent in NGS allows for high-throughput sequencing at low costs [6]. A range of various NGS technologies currently exist, from a number of different companies [7] (e.g., HiSeq, from Illumina; 454, from Roche Applied Science; Solid, from Applied Biosystems). These platforms are characterized by different approaches and can differ in terms of several technical specifications, such as sequencing cost per gigabyte (Gb), run time, reported accuracy, read length, observed raw error rate, sequence yield per run, insert size, instrument cost, and DNA requirements [8].
With this evolution in sequencing technologies, there has been ongoing progress in the field of genomics [9]. Hence, there has been an exponential increase in the use of various WGS applications in research and clinical practice [5], and it is expected to become a standard diagnostic tool in clinical practice [10, 11]. WGS has two general diagnostic potentials—namely, as a diagnostic instrument for manifested diseases [12] and as a predictive tool for determining disease dispositions [13, 14]. In many cases, WGS’s diagnostic and predictive potentials enhance patient benefits. In oncology, for example, a better understanding of cancer genetics, in tandem with improved disease diagnosis, prognosis, and management, can be achieved through the use of WGS. In the field of rare diseases, or in patients with an abnormal or an unknown phenotype, WGS may provide a diagnosis [15] and has the potential to end a diagnostic odyssey [16]. With a predictive approach, WGS may identify genetic variations, and predispositions to an increased risk for specific diseases [17]; for example, BRCA I and BRCA II are genetic mutations commonly linked to breast cancer [18]. Knowledge of various predispositions—as well as of incidental findings that are independent of previous diagnostic issues [19, 20] —can affect patient health through screening; it can also help mitigate risk and act as a part of various prevention measures [21]. Indeed, the results of WGS analyses can have far-reaching implications for patients [22]. The acquisition of genetic information can not only lead to behavioral changes in patients and their family members [23] but also increase the use of further diagnostics and of preventive and therapeutic procedures.
However, until recently, the diagnostic application of WGS was unthinkable, given its high procedure costs [24]. The cost of first decoding a human genome amounted to approximately US$3 billion [25]; even as of 2001, the cost of WGS was estimated at about US$100 million [26]. Meanwhile, technology firms yield at performing a WGS for less than US$1000 per genome [27–29]. However, the literature lacks relevant cost studies [30]. Additionally, it is necessary to consider and evaluate costs related to the clinical implementation of, and reimbursements for, undertaking WGS. Thus, in consideration of scarce resources and increasing expenses in the area of German healthcare, cost analyses in the run-up to WGS implementation as a diagnostic method are of significance. With this in mind, we conducted analyses of the costs of executing WGS, particularly in the context of German clinical practice.
Methodology
The creation of a standardized quality-assured process for WGS analysis, on the basis of procedures in the German Cancer Research Center (DKFZ), Heidelberg, constituted a starting point for the analysis described herein. The various steps within this process are defined with the help of expert opinions and clinical routines; thereafter, resources used in support of the process are identified. The overall costs per genome mainly depend on the applied sequencing platform used; hence, two sequencing platforms by DKFZ’s sequencing technology provider (i.e., Illumina, Inc.) were chosen. The first of these is the HiSeq 2500 (Illumina Inc.; San Diego, CA, USA), which is currently the standard device for high-throughput sequencing in most clinical facilities; the second—namely, the HiSeq Xten (Illumina Inc.; San Diego, CA, USA)—is the latest development in high-throughput sequencing, and it was studied to compare the effects of higher throughput.
General methodology
Step 1: Resource identification
Drawing on standard DKFZ processes, a quality-assured WGS process was generated. For this cost calculation, an institutional perspective was selected; indirect personnel costs were not calculated. Generally, single costs can be directly allocated to WGS, whereas while overhead costs are essential to the examination and organization of a WGS, they cannot be initially assigned to a single sequencing process. Hence, only direct medical costs and site-specific costs for sequencing devices essential to WGS execution were included; all other site-specific nonmedical direct costs and overhead costs (e.g., water, energy, administration expenses, and the use of IT infrastructure) were excluded from the analysis. Moreover, personnel costs were categorized as those pertaining to medical, technical, and bioinformatics personnel.
Step 2: Resource quantification
In the second step, the identified resources were quantified. It should be noted that complete utilization (i.e., 100 %) of the sequencing platforms is implausible, owing to maintenance, failures, cleaning, and missing sequencing assignments. Therefore, the effects of different utilization levels were analyzed, via sensitivity analysis. In this step, the influence of other levels of utilization (i.e., 90, 80, 70, and 60 %) on costs was simulated. Taking into account economies of scale and fixed-cost degression, the average costs of WGS were found to decrease with higher levels of utilization. Moreover, the depth of sequencing (coverage) is a substantial cost-influencing factor and correlates with error rate, amount of data generated, as well as the amount of genomes per run. In line with the desired level of accuracy, the coverage rate was chosen, and this rate influenced the amount of genomes per run; therefore, sensitivity analysis was undertaken with regards to various coverage values (i.e., 10×, 15×, 30×, 60×, and 75×). An increase in the average costs was found with increased coverage and the accompanying reduction in the number of genomes per run.
Step 3: resource evaluation
In this step, the identified and quantified resources were assessed in terms of monetary value. These monetary valuations were based on data and information provided by human genetic experts, hospitals, and private cooperation partners. Data used in the sequencing equipment and other materials were provided by Illumina, Inc., and their costs are based on the company’s list prices. The personnel working time for a single task was estimated using data from expert interviews. Subsequently, time estimations were valuated through the use of monetary mean values. Personnel costs for chemical–technical assistants (CTA) and bioinformaticians were calculated on the basis of the German civil service collective agreement of the federal state (TV–L) of Baden–Württemberg. Different pay-scale levels were used in these calculations: for bioinformaticians, a weekly working time of 39 h and an annual gross salary of €55,902.84 (€0.50 per minute) were assumed, and for CTAs, a weekly working time of 39 h and an annual gross salary of €40,809.33 (€0.36 per minute) were assumed. The payroll expenses for specialized clinical geneticists were based on the civil service collective agreement for physicians at the university clinics of the federal state of Baden–Württemberg; hence, a weekly working time of 39.30 h and an annual gross salary of €87,543.96 (€0.77 per minute) were assumed. For obtaining a blood sample, costs of €5.65—according to the uniform value scale, the basis of pricing of ambulant services (EBM)—were assumed. For an adequate calculation of the annual costs of acquisition and maintenance, we used the annuity method [31]. In this way, annual payments consisting of interest and redemption were calculated. For this purpose, an interest rate of 3 % was assumed.
Base case scenario
The cost of a single WGS analysis is influenced by several aspects, including the examination aim, clinical setting, technical aspects, data generation, and the use of sequencing platforms. In addition, depending on the aim of the examination, divergences emerge in diagnostic settings (in-patient vs. out-patient), scope of genetic counseling (general genetic screening vs. specific clinical issue), and genetic material acquisition (operation vs. blood test). For this reason, we defined a base case scenario. WGSs are typically performed when a rare disease is suspected. An out-patient setting in which genetic material is obtained via a blood sample is assumed. The clarification of secondary findings (e.g., according to the gene list of the American College of Medical Genetics and Genomics (ACMG)) was not included. Furthermore, the base case scenario is hallmarked by certain technical aspects, such as a sequencing platform utilization setting of 80 % and 30-times coverage.
Results
Process structure
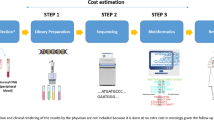
The cost analysis was based on the identification of relevant process steps. The process chart is illustrated in Fig. 1.
A three-step process structure was created that comprised pre-sequencing (direct patient contact and administration), sequencing (mechanical and biochemical processing of genetic material), and post-sequencing process (evaluation and final clinical genetic consultation).
Step 1: identification of necessary resources
The pre-sequencing process is characterized by direct patient contact. The first step prior to the diagnostic examination is patient administration. The pre- and post-sequencing clinical genetic consultation is, unlike research, an indispensable component of patient-centered quality management in clinical genetic care. The informed consent process, regarding opportunities and risks as well as the arrangement of relaying findings, is an important part of clinical genetic consultation. The time required for these medical consultations depends on the aim of the medical examination and the consequences of the patient results. In addition, genetic material is generally extracted from blood samples, smear tests, or during surgical interventions, so they incur costs at a variety of levels. The included costs of the pre-sequencing process comprise the personnel costs for the clinical geneticist and administrative employees, as well as the costs related to a blood sample.
During the sequencing process, the costs of diagnostic examinations emerge. The essential work steps are the mechanical and biochemical processing of genetic material, followed by the setting up of sequencing devices and cleaning. Additional costs in the sequencing process include personnel cost for technical staff, sequencing material and allocated costs for the acquisition, and maintenance of sequencing platforms.
The post-sequencing process is divided into the analysis, interpretation, and validation of acquired data, and final clinical genetic consultation, which includes conveying the findings to patients. In this step, the included personnel costs are those associated with clinical geneticists and bioinformaticians.
Steps 2 and 3: quantification and monetary valuation of resources
Costs of the pre-sequencing process
The pre-sequencing process is mainly characterized by personnel costs for initial clinical genetic consultation. With a time exposure of 45–60 min for this clinical genetic consultation, costs of €40.43 per WGS at an average time exposure of 52.5 min arise. Furthermore, costs of €5.65 for obtaining a blood sample were incurred; hence, the total cost for the pre-sequencing process amounts to €46.08.
Costs of sequencing process
The sequencing process costs consist of those for personnel such as technical staff and sequencing material, as well as costs allocated to the acquisition and maintenance of sequencing platforms.
Personnel costs
CTA personnel costs scarcely differ between the HiSeq 2500 and HiSeq Xten (Table 1). Moreover, it was found that the preparation of histoid is the most time-consuming step in the sequencing process, and it incurs personnel costs of €108.00.
Acquisition costs and maintenance costs
The acquisition costs of the sequencing platform on a per-genome basis amount to €485.29 for the HiSeq 2500, and €199.89 for the HiSeq Xten (Table 2). Despite the distinct lower acquisition costs associated with the HiSeq 2500, its higher per-genome cost emerges as a result of the time and quantity of genomes per run. This shows that the ‘time per run’ and the ‘number of sequenced genomes per run’ significantly influences overall costs. The operating life (which is synonymous with the technology life) is 3 years; given 80 % utilization, 30-times coverage, and a machine lifetime of 3 years, a maximum of 1458 human genomes can be sequenced with the HiSeq 2500, and 46,716 human genomes with the HiSeq Xten.
In addition, fixed costs for technical service and maintenance were found to be significant. Allocated costs for maintenance and service agreements amount to €122.11 for HiSeq 2500 and € 41.38 for HiSeq Xten per genome (Table 2).
Material costs
The costs associated with sequencing materials represent an essential cost factor, and they are split into 16 (per machine) and ten human genomes per run for the HiSeq Xten and HiSeq 2500, respectively. However, it should be noted that 160 genomes can be sequenced simultaneously on the HiSeq Xten. The material costs per run for sequencing with the HiSeq Xten are significantly higher than those with the HiSeq 2500. Nevertheless, dividing the material costs across a large number of analyses leads to significantly lower costs per genome for the HiSeq Xten (Table 3).
Sensitivity analysis of workload and coverage differentiation
The results of the two sensitivity analyses are shown in the appendix (Tables 5, 6). On account of larger economies of scale and fixed-cost degression, the average costs of a WGS analysis are reduced in relation to output quantity (Table 5). Assuming 80 % utilization, the total cost for materials, acquisition, and maintenance is €3455.48 and €1022.85 per genome for the HiSeq 2500 and HiSeq Xten, respectively. On the other hand, the costs per genome increase with an increase in coverage rate: a doubling of the coverage rate leads to a halving of the quantity of genomes per flowcell. For example, an increase in coverage from 30 times to 60 times reduces the number of genomes per run, from ten genomes to five on the HiSeq 2500. Hence, the costs associated with materials, acquisition, and maintenance increase to €6880.88 (Table 6).
Post-sequencing process costs
Personnel costs comprise an important cost factor in the post-sequencing process. These costs can be categorized as those for clinical geneticists and those for bioinformaticians. The mean cost of clinical geneticists is €40.43 for the final clinical genetic consultation, for an average time exposure of 52.5 min. Additional costs stem from work associated with bioinformatical interpretation; the duration of this task depends on the specific issue at hand, and can range from 1 h to a few days. However, in line with the base case scenario, six working hours was assumed. Hence, costs of €180.00 arise from undertaking a read-quality check (possible with read trimming), the identification of single nucleotide polymorphisms (SNP) or mutations, and the interpretation of identified SNPs or mutations.
Overall costs
Currently, the cost of a WGS analysis in a clinical setting in Germany is €3858.06 assuming 80 % utilization of the sequencing platform and a 30-times coverage with a HiSeq 2500. By using the latest high-throughput technology (i.e., HiSeq Xten), the overall cost could be reduced by 63 %, to €1411.20. The sequencing process—especially the sequencing materials and allocated investment costs—was identified as the most expensive WGS component. The results are summarized in Table 4.
Discussion
Currently, the cost of a WGS analysis in a clinical setting in Germany is €3858.06. To determine the costs of implementing this diagnostic procedure, evidence related to associated expenses is needed. In addition to medical evidence, cost evaluations are important to medical decision-makers; more importantly, especially for WGS, it is essential to determine which procedures will have a potentially high economic impact, and so reliable cost evaluations are necessary.
The overall cost of a WGS analysis depends on a plurality of aspects; in the following, the main cost-influencing factors—such as the sequencing platform used, the material costs, and the coverage rate—are highlighted. The selection of a high-throughput technology is the first major strategic decision in WGS implementation; this selection affects investment and maintenance expenses, as well as costs related to the sequencing materials that will be used. The results of this analysis showed that with a utilization rate of 80 % of the sequencing platform, the allocated acquisition costs of HiSeq Xten were about 60 % lower than those of HiSeq 2500, owing to higher throughput; the situation is similar for the costs of sequencing materials, which comprise the main cost factor in executing WGS analyses. Costs per genome are substantially influenced by the utilization of sequencing platforms and flowcells, as a result of the fixed-cost degression; hence, adopting the latest technology seems to be a precondition to keeping the average cost low. However, these circumstances need to be considered with caution. Keeping the average cost low assumes that a specific demand for genetic analysis, as well as a certain rate of utilization, is achieved. One HiSeq Xten has a significantly higher capacity (i.e., one HiSeq Xten can replace 32 HiSeq 2500 machines), and implementation may lead to significant overcapacity. Therefore, calculations that assume a utilization rate of 80 % for the HiSeq Xten might be overly high, and may therefore provide too-optimistic cost calculations. Lower utilization leads to an apportionment of fixed costs to fewer genomes, and thus to higher costs per genome. Hence, before a new sequencing platform is implemented, calculations of the probable number or future needs of WGS analysis during the operating time should be conducted. Moreover, future directions of the demand for WGS can scarcely be assessed at this time; this demand depends, for example, on national reimbursement regulations. The establishment of a limited number of WGS execution sites, perhaps in the form of centers, could possibly lead to the cost-effective execution of WGS; effective management and better utilization of sequencing platforms may help achieve these lower costs. However, genetic analyses are an emerging tool, and demand for its use will gradually increase. Therefore, technology firms should also look to develop platforms (e.g., HiSeq 4000 by Illumina, Inc.) with a higher utilization rate (relative to the HiSeq 2500) and lower acquisition costs (relative to the HiSeq Xten) to deal with what will no doubt be increasing demand, and to address the potential for significant overcapacity.
Test quality and costs correlate with the selection of coverage rate, which in turn influences the sensitivity of detection [35]. The selection of coverage rate is based on the intended validity of genetic analysis results. At this point, 30-times coverage is the customary benchmark for high-quality genome data [36]. However, with complex heterogenic genetic structures, the sequencing of tumors (for example) is largely conducted with significantly higher coverage rates [37, 38]. In general, the selection of coverage influences not only the number of genomes per run—and thus the costs per genome—but also (depending on the various amounts of genetic data) the cost of data storage and evaluation.
The highest personnel costs arise from bioinformatical work steps. The interpretation process is influenced by the purpose behind the examination, as well as the experience of the bioinformatician involved (these factors result in a wide range of time estimations, from 1 h to a few days). The time needed for data interpretation also depends on clinical issues, dataset size, and the hardware IT infrastructure being used. Costs associated with data validation—an additional cost-influencing work step—are difficult to calculate. Validations were conducted only if disease-relevant mutations are identified; these mutations and biomarkers were verified using traditional Sanger technology, which is the current ‘gold standard’ [39]. Conspicuous genetic features differ in frequency and by patient; however, an estimation of the frequency of specific conspicuous genetic features was not available. Hence, validation costs cannot be depicted.
Due to site specificity, cost-increasing factors—such as overhead and the costs of IT infrastructure and data storage—were excluded from the cost analysis. IT costs constitute a substantial part of investment costs, and long-term cooperation, quantity effects, and discounts determine these site-specific costs; they can also create substantial differences between list and project prices. Moreover, overhead—such as energy, water, rent, and administration costs—are characterized by high variability. Nevertheless, these costs should be considered in WGS reimbursement decisions.
Other limitations of this study include the single evaluation of WGS processes in DKFZ, the constraint of using a single technology, the fact that monetary evaluations are estimations made by clinical genetic experts, and the fact that data are from a single technology provider. The process chart is influenced by the specific structural organization of and processes in the DKFZ, and may differ with each institution or hospital. Investment, maintenance, and material costs were provided by Illumina, Inc.; these data were most appropriate in ensuring the study’s high representativeness, owing to the largest market share of the NGS market and therefore the worldwide distribution of sequencing platforms [40, 41]. It is noteworthy that the use of a sequencing platform from another provider may lead to different cost estimates.
An important finding of this study is that cost analyses for WGS, as an innovative diagnostic tool, cannot be generalized. Variations in relevant cost-influencing factors will necessarily lead to different overall costs. Therefore, the following aspects should be considered in any cost assessment of WGS: (1) the use of an inpatient versus outpatient setting, (2) the diagnostic context at hand (e.g., the costs of using WGS to inform cancer care differ greatly from those of using WGS to diagnose a rare disease, especially within the scope of genetic counselling), (3) the approach and technology used (e.g., different costs per Gb and time for sequencing), (4) the cost factors to be included (no consensus exists as to which cost factors should be included in a cost analysis of WGS), (5) the experience of the personnel involved (experience may influence processing time, including bioinformatical interpretations), and (6) regulations informed by secondary or incidental findings (e.g., confirmation with Sanger technology).
This study shows that, to date, the US$1000 genome has not become a reality in German quality-assured health care settings. However, technological progress may lead to further cost reductions, and so it may be possible to eventually achieve an even lower price for a single WGS [42]. The sole consideration in the development of costs for materials, acquisition, and maintenance—or the cost per Megabase of DNA Sequence [26]—suggests that, in the relatively near future, a US $1000 genome may become a reality. However, other process-relevant factors that ensure a quality WGS execution are both integral parts and fixed components of this process, and they are not subject to such cost reductions over time.
In addition to improvements to sequencing platforms, both databases and bioinformatics tools may be improved in the near future. Databases are prerequisite to genome-wide association studies. Databases are growing in size, and the body of knowledge on phenotype–genotype correlations will also steadily increase in size. Besides the increased body of genetic knowledge, improvements in bioinformatics tools will facilitate both faster and cheaper assessments of the pathogenicity of (novel) variants; in this way, faster and more precise diagnoses will be possible. These conditions offer considerable benefits in terms of patient care. Improvements in diagnosis, especially of diseases of an unknown phenotype, can also affect the cost-effectiveness of WGS; besides possible cost reductions, improvements in patient care (e.g., quality of life, time of diagnosis, and diagnosis and treatment options) may also lead to increased cost-effectiveness.
However, there are numerous ethical, legal, and economic barriers inherent in the unrestricted use of WGS. Given the predictive potential of using WGS, an increase in costs on account of incidental use is feared. Incidental findings may lead to further diagnostics, as well as preventive and therapeutic procedures. The development of these consequential follow-up costs—many of which are caused by behavioral changes in patients, physicians, and family members—cannot be assessed with certainty today. Hence, the widespread application of WGS should be rejected, and its use should be indicated only under certain conditions [15]. The unrestricted application of WGS, in tandem with a lack of limitations on feedback practices, will lead to an unquantifiable increase in healthcare expenses. Limitations on specific indications can prevent increases in expenditures. Therefore, defining the criteria by which WGS is indicated is a future responsibility for policy decision-makers. Furthermore, data security, the effects of genetic information on insurance policies and employment agreements, and the extent of insurance benefits are critical issues that relate to the application of WGS. In addition, certain regulations should be adopted prior to implementation [43–45].
Conclusions
The calculated cost of a single WGS was estimated at €3858.06 while assuming 80 % capacity utilization with the sequencing platform widely used in Germany. Although this study focused on medical costs, to derive a comprehensive illustration of costing in a quality-assured healthcare system, overhead should also be considered. Moreover, because of the high costs associated with a single WGS analysis, the application of this analysis should be limited to specific indications that promise substantial medical benefits for patients. Technical progress may lead to a further reduction in the cost of WGS analysis, and so the application of WGS in medical care as a diagnostic, predictive, and prognostic tool is most likely to become more widespread in future medical care.
References
Bentley, D.R., et al.: Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456(7218), 53–59 (2008). doi:10.1038/nature07517
Rockman, M.V.: Reverse engineering the genotype-phenotype map with natural genetic variation. Nature 456(7223), 738–744 (2008). doi:10.1038/nature07633
Shendure, J., Ji, H.: Next-generation DNA sequencing. Nat. Biotechnol. 26(10), 1135–1145 (2008). doi:10.1038/nbt1486
Sanger, F., Nicklen, S., Coulson, A.R.: DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74(12), 5463–5467 (1977)
Renkema, K.Y., Stokman, M.F., Giles, R.H.: Knoers, Nine V A M: next-generation sequencing for research and diagnostics in kidney disease. Nat. Rev. Nephrol. 10(8), 433–444 (2014). doi:10.1038/nrneph.2014.95
Grada, A., Weinbrecht, K.: Next-generation sequencing: methodology and application. J. Invest. Dermatol. 133(8), e11 (2013). doi:10.1038/jid.2013.248
Buermans, H.P.J., den Dunnen, J.T.: Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta 1842(10), 1932–1941 (2014). doi:10.1016/j.bbadis.2014.06.015
Quail, M.A., Smith, M., Coupland, P., Otto, T.D., Harris, S.R., Connor, T.R., Bertoni, A., Swerdlow, H.P., Gu, Y.: A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 13, 341 (2012). doi:10.1186/1471-2164-13-341
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L., Law, M.: Comparison of next-generation sequencing systems. J Biomed. Biotechnol 2012, 251364 (2012). doi:10.1155/2012/251364
Green, E.D., Guyer, M.S.: Charting a course for genomic medicine from base pairs to bedside. Nature 470(7333), 204–213 (2011). doi:10.1038/nature09764
Kingsmore, S.F., Saunders, C.J.: Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Science translational medicine 3(87), 87ps23 (2011). doi:10.1126/scitranslmed.3002695
Choi, M., Scholl, U.I., Ji, W., Liu, T., Tikhonova, I.R., Zumbo, P., Nayir, A., Bakkaloğlu, A., Ozen, S., Sanjad, S., Nelson-Williams, C., Farhi, A., Mane, S., Lifton, R.P.: Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 106(45), 19096–19101 (2009). doi:10.1073/pnas.0910672106
Carlson, C.S., Eberle, M.A., Kruglyak, L., Nickerson, D.A.: Mapping complex disease loci in whole-genome association studies. Nature 429(6990), 446–452 (2004). doi:10.1038/nature02623
Meuwissen, T., Goddard, M.: Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185(2), 623–631 (2010). doi:10.1534/genetics.110.116590
ACMG Board of Directors: Points to consider in the clinical application of genomic sequencing. Genetics Med Off J Am Coll Med Gen 14(8), 759–761 (2012). doi:10.1038/gim.2012.74
Sawyer, S.L., Hartley, T., Dyment, D.A., Beaulieu, C.L., Schwartzentruber, J., Smith, A., Bedford, H.M., Bernard, G., Bernier, F.P., Brais, B., Bulman, D.E., Warman Chardon, J., Chitayat, D., Deladoëy, J., Fernandez, B.A., Frosk, P., Geraghty, M.T., Gerull, B., Gibson, W., Gow, R.M., Graham, G.E., Green, J.S., Heon, E., Horvath, G., Innes, A.M., Jabado, N., Kim, R.H., Koenekoop, R.K., Khan, A., Lehmann, O.J., Mendoza-Londono, R., Michaud, J.L., Nikkel, S.M., Penney, L.S., Polychronakos, C., Richer, J., Rouleau, G.A., Samuels, M.E., Siu, V.M., Suchowersky, O., Tarnopolsky, M.A., Yoon, G., Zahir, F.R., Majewski, J., Boycott, K.M.: Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Gen. (2015). doi:10.1111/cge.12654
Berg, J.S., Khoury, M.J., Evans, J.P.: Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genetics Med. 6, 499–504 (2011). doi:10.1097/GIM.0b013e318220aaba
Ford, D., Easton, D.F., Stratton, M., Narod, S., Goldgar, D., Devilee, P., Bishop, D.T., Weber, B., Lenoir, G., Chang-Claude, J., Sobol, H., Teare, M.D., Struewing, J., Arason, A., Scherneck, S., Peto, J., Rebbeck, T.R., Tonin, P., Neuhausen, S., Barkardottir, R., Eyfjord, J., Lynch, H., Ponder, B., Gayther, S.A., Birch, J.M., Lindblom, A., Stoppa-Lyonnet, D., Bignon, Y., Borg, A., Hamann, U., Haites, N., Scott, R.J., Maugard, C.M., Vasen, H., Seitz, S., Cannon-Albright, L.A., Schofield, A., Zelada-Hedman, M.: Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am. J. Human Gen. 62(3), 676–689 (1998). doi:10.1086/301749
Green, R.C., Berg, J.S., Grody, W.W., Kalia, S.S., Korf, B.R., Martin, C.L., McGuire, A.L., Nussbaum, R.L., O’Daniel, J.M., Ormond, K.E., Rehm, H.L., Watson, M.S., Williams, M.S., Biesecker, L.G.: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Gen. Med. Off. J. Am. Coll. Med. Gen. 15(7), 565–574 (2013). doi:10.1038/gim.2013.73
Knoppers, B.M., Deschênes, M., Zawati, M.H., Tassé, A.M.: Population studies: return of research results and incidental findings policy statement. Eur. J. Human Gen. EJHG 21(3), 245–247 (2013). doi:10.1038/ejhg.2012.152
Evans, J.P., Skrzynia, C., Burke, W.: The complexities of predictive genetic testing. BMJ 322(7293), 1052–1056 (2001)
Dondorp, W.J., de Wert, W.J., Guido, M.W.R.: The ‘thousand-dollar genome’: an ethical exploration. Eur. J. Hum. Genet. 21(Suppl 1), S6–26 (2013). doi:10.1038/ejhg.2013.73
McBride, C.M., Koehly, L.M., Sanderson, S.C., Kaphingst, K.A.: The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annu. Rev. Public Health 31, 89–103 (2010). doi:10.1146/annurev.publhealth.012809.103532
Ten Bosch, John, R., Grody, W.W.: Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J Mol. Diagn JMD 10(6), 484–492 (2008). doi:10.2353/jmoldx.2008.080027
National Human Genome Research Institute: The Human Genome Project Completion: Frequently Asked Questions (2010). https://www.genome.gov/11006943. Assessed 17 July 2015
Wetterstrand, K.A.: DNA sequencing costs: data from the NHGRI Genome Sequencing Program. http://www.genome.gov/sequencingcosts. Assessed 13 January 2015
Collins, F.: Has the revolution arrived? Nature 464(7289), 674–675 (2010). doi:10.1038/464674a
Hayden, E.C.: Technology: the $1,000 genome nature. Nature 507(7492), 294–295 (2014). doi:10.1038/507294a
Service, R.F.: Gene sequencing. The race for the $1000 genome. Science (New York, N.Y.) 311(5767), 1544–1546 (2006). doi:10.1126/science.311.5767.1544
Frank, M., Prenzler, A., Eils, R., Graf von der Schulenburg, J-M.: Genome sequencing: a systematic review of health economic evidence. Health economics review 3(1), 29 (2013). doi: 10.1186/2191-1991-3-29
Götze, U., Northcott, D., Schuster, P.: Investment appraisal. Methods and models. Springer texts in business and economics
Illumina, Inc.: cBot (2015). http://www.illumina.com/products/cbot.html. Assessed 17 March 2015
Holt, R.A., Jones, Steven J M: The new paradigm of flow cell sequencing. Genome research 18(6), 839–846 (2008). doi:10.1101/gr.073262.107
QIAGEN: QIAamp DNA Blood Mini Kit (2016). https://www.qiagen.com/de/shop/sample-technologies/dna/dna-preparation/qiaamp-dna-blood-mini-kit#orderinginformation. Assessed 01 March 2016
Meynert, A.M., Ansari, M., FitzPatrick, D.R., Taylor, M.S.: Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinfo. 15, 247 (2014). doi:10.1186/1471-2105-15-247
Dewey, F.E., Pan, S., Wheeler, M.T., Quake, S.R., Ashley, E.A.: DNA sequencing: clinical applications of new DNA sequencing technologies. Circulation 125(7), 931–944 (2012). doi:10.1161/CIRCULATIONAHA.110.972828
Schlötterer, C., Tobler, R., Kofler, R., Nolte, V.: Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15(11), 749–763 (2014). doi:10.1038/nrg3803
Kofler, R., Schlötterer, C.: A guide for the design of evolve and resequencing studies. Mol. Biol. Evol. 31(2), 474–483 (2014). doi:10.1093/molbev/mst221
McCourt, C.M., McArt, D.G., Mills, K., Catherwood, M.A., Maxwell, P., Waugh, D.J., Hamilton, P., O’Sullivan, J.M., Salto-Tellez, M.: Validation of next generation sequencing technologies in comparison to current diagnostic gold standards for BRAF, EGFR and KRAS mutational analysis. PLoS One 8(7), e69604 (2013). doi:10.1371/journal.pone.0069604
Karow, J.: In Sequence 2013 Survey: Illumina Pulls Further Ahead, Interest in Oxford Nanopore Remains High (2014)
Studt, T,: Innovations Drive Rapid NGS Growth. Lower costs, high throughput and enhanced accuracies provide next-generation sequencing users with enhanced medical information. (2015)
PHG Foundation: PHG Foundation: Next steps in the sequence. The implications of whole genome sequencing for health in the UK, Cambridge (2011). http://www.phgfoundation.org/file/10363/. Assessed 23 April 2015
National Human Genome Research Institute: Genetic Discrimination (2014). http://www.genome.gov/10002077. Accessed 23 April 2015
American Society of Clinical Oncology policy statement update: Genetic testing for cancer susceptibility. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 21(12), 2397–2406 (2003). doi:10.1200/JCO.2003.03.189
Statement of the ESHG on direct-to-consumer genetic testing for health-related purposes. European journal of human genetics: EJHG 18(12), 1271–1273 (2010). doi:10.1038/ejhg.2010.129
Acknowledgments
We thank the German Cancer Research Center (DKFZ), Heidelberg; Illumina Inc., San Diego, CA, USA; Hanover Medical School (MHH); German Faculty of Mathematics and Computer Science Chair of RNA Bioinformatics and High Throughput Analysis at the Friedrich-Schiller-University, Jena; Max Planck Institute, Berlin; Center for Human Bioinformatics, Heidelberg; Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB, Stuttgart; Bernhard Nocht Institute for Tropical Medicine, Hamburg; Helmholtz Centre, Munich; Bioinformatics and System Biology Justus Department at the Liebig-University Gießen; and the Institute for Biochemistry, Biotechnology and Bioinformatics at the Technical University of Braunschweig and BioNTech RNA Pharmaceuticals GmbH for their valuable information.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Plöthner, M., Frank, M. & von der Schulenburg, JM.G. Cost analysis of whole genome sequencing in German clinical practice. Eur J Health Econ 18, 623–633 (2017). https://doi.org/10.1007/s10198-016-0815-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10198-016-0815-0