Abstract
Accurate determination of resistance is important to ensure appropriate antimicrobial therapy in Stenotrophomonas maltophilia infections. This study was undertaken to evaluate the susceptibility results obtained by disc diffusion, E-test, Phoenix system, and reference agar dilution method and also to evaluate the in vitro activity of various antimicrobial combinations against multidrug-resistant S. maltophilia. Susceptibilities to several antimicrobial agents were determined by agar dilution, disc diffusion, and E-test according to the US Clinical Laboratory and Standards Institute (CLSI) guidelines. Results were also evaluated in the in Phoenix system for available agents. Twelve different antibiotic combinations were tested for synergy by the E-test method. Most synergic combinations were confirmed by microdilution checkerboard assay. Tigecycline, trimethoprim/sulfamethoxazole (TMP–SMX) and doxycycline were the most effective drugs against S. maltophilia. Poorest agreement was determined by disc diffusion and E-test against ticarcillin/clavulanate and ciprofloxacin (κ < 0.4), by disc diffusion against colistin (κ < 0.4), and by the Phoenix system against piperacillin/tazobactam (κ < 0.4). Based on these data, disc diffusion seems to be unreliable for ticarcillin/clavulanate, ciprofloxacin, and colistin; E-test for ticarcillin/clavulanate and ciprofloxacin; and the Phoenix system for piperacillin/tazobactam for S. maltophilia susceptibility testing. Synergistic activity was detected predominantly with TMP–SMX + ticarcillin/clavulanate and TMP–SMX + ceftazidime. TMP–SMX + ceftazidime synergy was also supported by the checkerboard method. However, TMP–SMX + ticarcillin/clavulanate combination revealed indifferent effect by the checkerboard assay. As ticarcillin/clavulanate and ciprofloxacin E-test results were beyond the acceptable correlation limits, synergy testing performed with these agents was considered as unreliable. Further studies are required to standardize susceptibility testing, especially for colistin, ticarcillin/clavulanate, and ciprofloxacin for S. maltophilia. TMP–SMX-containing drug combinations seemed to be more synergistic on multidrug-resistant S. maltophilia; however, these results merit further evaluation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stenotrophomonas maltophilia has emerged as an opportunistic nosocomial pathogen of increasing importance. As it is intrinsically resistant to a broad spectrum of antimicrobial agents, the treatment of infections due to S. maltophilia is usually problematic in clinical practice, leading to a high frequency of treatment failure and mortality [1, 2]. The antimicrobial resistance of S. maltophilia is attributed to the reduction in outer membrane permeability, expression of efflux pumps, or production of multiple beta-lactamases [3, 4]. The expression of two or more of these resistance mechanisms together usually results in the development of multidrug resistance (MDR), a condition that may necessitate the use of antimicrobial agents in combination. Antimicrobial susceptibility testing of S. maltophilia isolates presents some problems, and susceptibility testing guidelines have not yet been fully established for this microorganism. The US Clinical Laboratory and Standards Institute (CLSI) recommends the use of the broth or agar dilution method and disc diffusion method for testing minocycline, levofloxacin, and trimethoprim/sulfamethoxazole (TMP–SMX); and the broth or agar dilution method for testing ticarcillin/clavulanic acid, ceftazidime, and chloramphenicol [5]. Besides standard manual methods, several automated systems are now available for identification and susceptibility testing of bacteria. However, various drawbacks have been reported about the accuracy and reliability of such systems in the determination of antimicrobial susceptibilities [6–8]. This study was conducted to evaluate and compare the methods of agar dilution, disc diffusion, the E-test, and the BD Phoenix automated susceptibility testing system to determine the susceptibility profile of S. maltophilia to nine different antimicrobial agents and also to evaluate the potential of various antimicrobial combinations against MDR S. maltophilia.
Materials and methods
Isolates
A total of 25 nonduplicate, genetically unrelated S. maltophilia strains isolated from individual patients hospitalized at Hacettepe University Hospital, Ankara, Turkey, in 2005 were included to the study. Identification of the isolates was carried out by BD Phoenix Automated Identification and Susceptibility Testing System (Becton–Dickinson Biosciences, USA). Identification of the isolates was confirmed by standard microbiological tests such as Gram stain, oxidase, DNase, and lysine decarboxylase [9]. Genetic unrelatedness of the isolates was previously proven by pulsed-field gel electrophoresis [10]. The band patterns were interpreted according to the criteria of Tenover et al. [11], with patterns that differed by two or three bands being defined as closely related subtypes.
Susceptibility testing
The tested antimicrobial agents were ceftazidime, piperacillin/tazobactam, ticarcillin/clavulanic acid, meropenem, TMP–SMX, ciprofloxacin, doxycycline, tigecycline, and colistin. Disc diffusion, agar dilution, and E-test methods were applied to determine susceptibilities of the test isolates. The antimicrobial discs were obtained from BBL, Becton–Dickinson. The drug powders were supplied by their manufacturers or obtained commercially. Disc diffusion and agar dilution were performed according to CLSI guidelines [12]. The E-test (AB Biodisk, Solna, Sweden) method was applied according to the manufacturer’s instructions. Mueller–Hinton agar (Acumedia, USA) was used as the susceptibility test medium, and all test media were incubated at 35°C in normal atmosphere for 20–24 h. Escherichia coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as quality-control strains. The susceptibility test results were interpreted according to CLSI criteria if susceptibility breakpoints existed for the specific agent [12]. Otherwise, the results were evaluated using CLSI criteria for P. aeruginosa, Acinetobacter spp. or non-Enterobacteriaceae other than P. aeruginosa [12]. As tigecycline breakpoints have not been established as yet, tigecycline results were interpreted according to breakpoints reported in the literature as ≤2, 4, and ≥8 μg/ml for susceptible, intermediate, and resistant isolates, respectively [13, 14]. Isolates resistant to more than two classes of antimicrobial agents were defined as MDR. Isolates in the intermediate susceptibility category were accepted as resistant for the calculation of resistance rates. Susceptibility tests with the BD Phoenix system were performed using panel NMIC/ID-55 or NMIC/ID-5 for gram-negative rods (ref. no. 448935) according to the manufacturer’s recommendations. Phoenix results were obtained only for ceftazidime, piperacillin/tazobactam, meropenem, and TMP–SMX.
Synergy testing
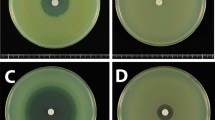
For synergy testing, 12 different combinations of E-test strips were placed on Mueller–Hinton agar medium in a cross formation, with a 90° angle at the intersection between the scales at the respective minimal inhibitory concentrations (MICs) for S. maltophilia, and the plates were incubated at 35°C for 24 h (Fig. 1) [15]. The MICs were interpreted at the point of intersection between the inhibition zone and the E-test strip. The fractional inhibitory concentration index (ΣFIC) was calculated on the basis of the resultant zone of inhibition, as follows: ΣFIC = FIC A + FIC B, where FIC A is the MIC of the combination/MIC of drug A alone, and FIC B is the MIC of the combination/MIC of drug B alone. Synergy was defined by a ΣFIC of ≤0.5. Antagonism was defined by ΣFIC of >4. ΣFIC of >0.5 but ≤4 was interpreted as indifferent [16]. Two different combinations indicating the highest rate of synergy were further studied by microdilution checkerboard method for to confirm the E-test synergy results. These two combinations were chosen among the combinations containing drugs with the least reliable E-test susceptibility results.
Statistical analysis
Categorical agreement was used to compare susceptibilities obtained with disc diffusion, E-test, and BD Phoenix system with those obtained by the reference agar dilution method. Errors were determined by methods published in the National Committee for Clinical Laboratory Standards (NCCLS) M23-A2 and Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems; Guidance for Industry and FDA [17, 18]. Very major errors (VME) were considered when the organism resistant by the reference method was interpreted as susceptible by the test method. Major errors (MEs) were defined when the organism susceptible by the reference method was interpreted as resistant by the test method. Minor errors (mE) were considered when the reference method yielded a resistant or susceptible result but the test method yielded an intermediate result and vice versa. Kappa (κ) measure of agreement was used to define the correlation between tests; κ < 0.4 were interpreted as poor agreement, κ = 0.4–0.75 as fair to good agreement, and κ > 0.75 as excellent agreement.
Results
Twenty-five S. maltophilia strains were selected from a collection of S. maltophilia isolates previously shown to be genetically unrelated by pulsed-field gel electrophoresis [10]. Resistance rates determined for the nine antimicrobial agents against S. maltophilia obtained by reference agar dilution, disc diffusion, E-test, and BD Phoenix systems are given in Table 1. MDR was detected in 96% of isolates. Higher rates of susceptibility were detected for colistin by disc diffusion. No resistance was detected against tigecycline with any of the test methods. TMP–SMX and doxycycline were the following most effective drugs against S. maltophilia.
A total of 575 results were obtained when 25 S. maltophilia were tested against nine drugs (Phoenix system involved only five drugs) by three different testing methods. Categorical analysis of results revealed an overall category error rate of 21.3% for disc diffusion, with 27/225 (12%) of these being mE, 6/225 (2.7%) ME, and 15/225 (6.7%) VME; an overall categorical error rate of 23.6% for the E-test, with 40/225 (17.8%) being mE, 3/225 (1.3%) being ME, and 10/225 (4.4%) being VME; and an overall categorical error rate of 13.6% for the Phoenix system, with 10/125 (8%) being mE, 2/125 (1.6) being ME, and 5/125 (4%) being VME (Table 2). MEs were detected mainly for ticarcillin/clavulanate by disc diffusion and E-test. VMEs were determined for ceftazidime and ciprofloxacin by three methods and for piperacillin/tazobactam by the Phoenix system only. Colistin results obtained by disc diffusion revealed a high rate of VMEs (32%). High VME rates were also obtained for ciprofloxacin by disc diffusion (24%) and E-test (32%). Poorest agreement was determined by disc diffusion against ticarcillin/clavulanate, colistin, and ciprofloxacin (κ < 0.4), by E-test against ticarcillin/clavulanate and ciprofloxacin (κ < 0.4), and by the Phoenix system against piperacillin/tazobactam (κ < 0.4). Fair to good agreement was assessed for the E-test with ceftazidime and colistin and for the Phoenix system with TMP–SMX (κ = 0.64). All other agents and methods showed excellent agreement with the reference agar dilution method (κ > 0.75).
Synergistic activity was detected predominantly with TMP–SMX plus ticarcillin/clavulanate and TMP–SMX plus ceftazidime (Table 3). Colistin plus TMP–SMX and colistin plus tigecycline yielded antagonism in one isolate. Although for certain combinations MIC values of one of the drugs in combination decreased even below susceptibility breakpoints, ΣFIC indices were not always in the synergy range. However, in a certain number of such isolates, combinations including tigecycline, TMP–SMX, or doxycycline yielded significantly decreased MIC values for colistin or ceftazidime (Table 4).
As TMP–SMX plus ceftazidime and TMP–SMX plus ticarcillin/clavulanate indicated the highest rates of synergistic activity, these combinations were further tested by the microdilution checkerboard method to confirm the results. Whereas TMP–SMX plus ceftazidime revealed synergy in 72% of isolates by the E-test method, this rate was 56% by the checkerboard method. For TMP–SMX plus ticarcillin/clavulanate, whereas the E-test detected synergy in 64% of isolates, checkerboard detected only indifferent effects for all isolates.
Discussion
Resistance of S. maltophilia to several antimicrobial agents restricts the choice of drugs for treating such infections. MDR strains further complicate the problem, as empiric antibiotic therapy usually remains inadequate or inappropriate in such patients. In clinical practice, TMP–SMX, which has relatively low rates of resistance, has become the treatment of choice in S. maltophilia infections [19, 20]. The results of this study indicated high levels of MDR in S. maltophilia isolates, 96% of them being resistant to beta-lactam agents, ciprofloxacin, and colistin. The most active agents were tigecycline, doxycycline, and TMP–SMX, in accordance with previous data. There have been increasing reports of TMP–SMX resistance among S. maltophilia strains; however, in this study and other studies from Turkey, TMP–SMX resistance rates for S. maltophilia still remain low [10, 21]. Tigecycline, which has been shown to be one of the most promising therapeutic options for treating MDR Acinetobacter spp. and S. maltophilia infections [14, 22], has proven its good in vitro activity against S. maltophilia in this study, as well. One strain resistant to TMP–SMX and doxycycline was susceptible only to tigecycline.
Empiric therapeutic drugs may be switched to others after obtaining antibiotic susceptibility profiles specific for the causative agent. Thus reliable and accurate susceptibility testing is of crucial importance in the setting of S. maltophilia infections, which usually occur in immunocompromised patients or patients in intensive care units and necessitate prompt and appropriate antimicrobial therapy. CLSI has documented susceptibility testing guidelines for S. maltophilia against certain drugs, including ticarcillin/clavulanate, ceftazidime, minocycline, levofloxacin, TMP–SMX, and chloramphenicol [5]. The reference susceptibility testing is considered as agar dilution, and this study aimed to compare different testing methods for S. maltophilia and evaluate the results under the light of agar dilution MICs.
In general, disc diffusion or automated susceptibility testing systems are more preferred nowadays by the clinical microbiology laboratories for susceptibility testing, as dilution methods are labor intensive and have a higher cost. However, as results of this study also revealed, there are discrepant susceptibility data, particularly with disc diffusion for colistin, ticarcillin/clavulanate, and ciprofloxacin and with the E-test for ticarcillin/clavulanate, ciprofloxacin and, to a lesser extent, ceftazidime. There have been several reports related to the discordance between different susceptibility testing methods for S. maltophilia [6–8, 23–25]. It has been previously reported by the NCCLS that for an acceptable performance of susceptibility tests, the overall categorical error rate should be <10%, of these ≤1.5% being VMEs and ≤3% being MEs [17]. Although most discrepancies in this study were in the form of mEs, colistin and ticarcillin/clavulanate disc diffusion results exhibited very poor agreement with the reference method. The high VME rate for colistin obtained by the disc diffusion method is in accordance with previous reports, confirming the unacceptable accuracy and reliability of disc diffusion for testing colistin [26, 27]. Colistin results obtained with the E-test were within acceptable ranges, with most discrepancies being in the category of mEs. Thus, the E-test can be considered a good alternative for colistin MIC determination in S. maltophilia isolates. Our results with colistin in the E-test were inconsistent with the results of Tan et al. [26], who reported unacceptable error rates for the colistin E-test in S. maltophilia and P. aeruginosa. However, our data were in accordance with Nicodemo et al., reporting 96.7% essential agreement and Galani et al. reporting no VMEs or MEs between the E-test and agar dilution for colistin [27, 28]. When ciprofloxacin and ticarcillin/clavulanate results were evaluated, it was observed that disc diffusion and the E-test had poor agreement, with high VME rates, whereas Phoenix exhibited fair to good agreement. Poor agreement for ciprofloxacin and ticarcillin/clavulanate by disc diffusion and the E-test was also reported by several other authors [23–25]. These previously reported results were similar to ours in that the E-test and disc diffusion revealed false susceptibility to ciprofloxacin when compared with reference agar dilution. The possibilities that ciprofloxacin stability was poor or that degradation occurred could not be ruled out with the current data. However, ciprofloxacin MIC values were within normal ranges for the quality-control strains used in each run. Thus, lack of correlation of ciprofloxacin results in this study may be related to factors specific to S. maltophilia and quinolone resistance mechanisms. When ticarcillin/clavulanate results were evaluated, it was observed that more resistance was detected by the E-test, probably due to the presence of hazy colony growths within the inhibition ellipses.
When the overall categorical error rates were considered, disk diffusion and the E-test seemed to have similar rates of about 21–23%. Although this rate was higher than the acceptable rate, when examined in detail, serious discords were not detected, and it can be stated that E-test reliably carries out susceptibility testing of most of the antimicrobial agents included in the study, as VME and ME rates were within acceptable limits—except for ciprofloxacin, ticarcillin/clavulanate and, to a lesser extent, ceftazidime. As the Phoenix system gave susceptibility results for a limited number of agents, in this study, only ceftazidime, piperacillin/tazobactam, meropenem, TMP–SMX, and ciprofloxacin data were compared. According to this comparison, inconsistent results were obtained mainly for piperacillin/tazobactam, ceftazidime, and ciprofloxacin and also in a single isolate for TMP–SMX. The overall categorical error rates revealed that the Phoenix system has some inadequacies in susceptibility testing for S. maltophilia, especially with beta-lactams. Discrepant results with ceftazidime, cefotaxime, cefepime, ciprofloxacin, and TMP–SMX were also reported by Stefaniuk et al. for the BD Phoenix system [8]. Although it is still a matter of debate as to treatment guidance according to MIC values, it could be recommended that Phoenix susceptibility results may be applied cautiously for that purpose in case of S. maltophilia isolates.
A systematic literature review revealed that when cotrimoxazole administration is not possible, ciprofloxacin and ceftazidime or ceftriaxone administered as monotherapy or in combination with other agents had clinical success rates ranging from 66.7% to 85% in the limited number of reported cases [29]. However, S. maltophilia has resistance mechanisms for both of these classes of agents. Although the choice of monotherapy or combination therapy is a controversial issue, several authors suggest combination treatment, especially in patients at risk [29–31]. Therefore, synergy testing may help determine the most appropriate combination in each special setting. Synergy testing in this study revealed that the most effective antibiotic combinations against S. maltophilia were TMP–SMX plus ceftazidime, followed by TMP–SMX plus ticarcillin/clavulanate. Poulos et al. [30] reported TMP–SMX plus ticarcillin/clavulanate synergy for all S. maltophilia isolates tested. Liaw also suggested TMP–SMX plus ticarcillin/clavulanate and ceftazidime plus ciprofloxacin as the most synergic combinations against S. maltophilia isolates [31]. Colistin combined with TMP–SMX or doxycycline exhibited synergy in 36% of the isolates. Although this rate obtained according to ΣFIC indices does not seem to be satisfactory in terms of synergy, it is of note that most isolates showed colistin MIC values were lowered in combination with TMP–SMX, tigecycline, and doxycycline. Ciprofloxacin-added combinations are usually recommended for S. maltophilia infections [29]. However, as ciprofloxacin E-test results revealed poor agreement in this study, and synergy testing was performed by the E-test, ciprofloxacin synergy results did not seem to be reliable. Confirmation of the E-test synergy results by microdilution checkerboard method revealed agreement for TMP–SMX plus ceftazidime combination. However, results for the TMP–SMX plus ticarcillin/clavulanate combination were all in the category of indifferent effect by the checkerboard method. This could be attributed to the poorest agreement detected for ticarcillin/clavulanate by the E-test. Thus, E-test synergy results seem to be unreliable for ticarcillin/clavulanate also. The net effect of antimicrobial combinations against S. maltophilia should further be evaluated in both the animal model and the clinical setting. It is noteworthy that in vitro studies lack the effects of host immune response. Nevertheless, antibiotic combinations may provide significant benefit over monotherapy in MDR pathogens such as S. maltophilia.
References
Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:57–80.
Gales AC, Jones RN, Forward KR, Linares J, Sader HS, Verhoef J. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997–1999). Clin Infect Dis. 2001;32(Suppl 2):S104–13.
Zhang L, Li XZ, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother. 2000;44:287–93.
Li XZ, Zhang L, Poole K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2002;46:333–43.
CLSI. Performance standards for antimicrobial susceptibility testing. Nineteenth information supplement, CLSI document M100-S19. Wayne, PA: Clinical Laboratory Standards; 2009.
Carroll KC, Cohen S, Nelson R, Campbell DM, Claridge JD, Garrison MW, et al. Comparison of various in vitro susceptibility methods for testing Stenotrophomonas maltophilia. Diagn Microbiol Infect Dis. 1998;32:229–35.
Joyanes P, del Carmen Conejo M, Martinez-Martinez L, Perea EJ. Evaluation of the VITEK 2 system for the identification and susceptibility testing of three species of nonfermenting gram-negative rods frequently isolated from clinical samples. J Clin Microbiol. 2001;39:3247–53.
Stefaniuk E, Baraniak A, Gniadkowski M, Hryniewicz W. Evaluation of the BD Phoenix automated identification and susceptibility testing system in clinical microbiology laboratory practice. Eur J Clin Microbiol Infect Dis. 2003;22:479–85.
Winn W Jr, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P, Woods G. Koneman’s color atlas and textbook of diagnostic microbiology. 5th ed. Philadelphia: Lippinncott Williams & Wilkins; 2006.
Gulmez D, Hascelik G. Stenotrophomonas maltophilia: antimicrobial resistance and molecular typing of an emerging pathogen in a Turkish university hospital. Clin Microbiol Infect. 2005;11:880–6.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
CLSI. Performance standards for antimicrobial susceptibility testing. Seventeenth information supplement. CLSI Document M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute; 2007.
Pachon-Ibanez ME, Jimenez-Mejias ME, Pichardo C, Llanos AC, Pachon J. Activity of tigecycline (GAR-936) against Acinetobacter baumannii strains, including those resistant to imipenem. Antimicrob Agents Chemother. 2004;48:4479–81.
Insa R, Cercenado E, Goyanes MJ, Morente A, Bouza E. In vitro activity of tigecycline against clinical isolates of Acinetobacter baumannii and Stenotrophomonas maltophilia. J Antimicrob Chemother. 2007;59:583–5.
BD White RL, Manduru M, Bosso JA. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard and E-test. Antimicrobial Agents Chemother. 1996;40:1914–8.
Satish KP, MR, Eliopulos GM. Antimicrobial combinations. In: Lorien V, editor. Antibiotics in laboratory medicine. Philadelphia: Lippincott Williams and Wilkins; 2005. p. 365–440.
National Committee for Clinical Laboratory Standards N. Development of ın vitro susceptibility testing criteria and quality control parameters: approved standard M23-A2. Wayne: NCCLS; 2000.
FDA. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems; guidance for ındustry and FDA. US Department of Health and Human Services Food and Drug Administration Center for Devices and Radiological Health; 2007.
Betriu C, Sanchez A, Palau ML, Gomez M, Picazo JJ. Antibiotic resistance surveillance of Stenotrophomonas maltophilia, 1993–1999. J Antimicrob Chemother. 2001;48:152–4.
Valdezate S, Vindel A, Loza E, Baquero F, Canton R. Antimicrobial susceptibilities of unique Stenotrophomonas maltophilia clinical strains. Antimicrob Agents Chemother. 2001;45:1581–4.
Koseoglu O, Sener B, Gulmez D, Altun B, Gur D. Stenotrophomonas maltophilia as a nosocomial pathogen. New Microbiol. 2004;27:273–9.
Hoban DJ, Bouchillon SK, Johnson BM, Johnson JL, Dowzicky MJ. In vitro activity of tigecycline against 6792 gram-negative and gram-positive clinical isolates from the global tigecycline evaluation and surveillance trial (TEST Program, 2004). Diagn Microbiol Infect Dis. 2005;52:215–27.
JM Pankuch GA, Rittenhouse SF, Appelbaum PC. Susceptibilities of 123 strains of Xanthomonas maltophilia to eight beta-lactams (including beta-lactam-beta-lactamase inhibitor combinations) and ciprofloxacin tested by five methods. Antimicrobial Agents Chemother. 1994;38:2317–22.
Tatman-Otkun M, Gurcan S, Ozer B, Aydoslu B, Bukavaz S. The antimicrobial susceptibility of Stenotrophomonas maltophilia isolates using three different methods and their genetic relatedness. BMC Microbiol. 2005;5:24.
Krueger TS, Clark EA, Nix DE. In vitro susceptibility of Stenotrophomonas maltophilia to various antimicrobial combinations. Diagn Microbiol Infect Dis. 2001;41:71–8.
Tan TY, Ng SY. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect. 2007;13:541–4.
Galani I, Kontopidou F, Souli M, Rekatsina PD, Koratzanis E, Deliolanis J, et al. Colistin susceptibility testing by Etest and disk diffusion methods. Int J Antimicrob Agents. 2008;31:434–9.
Nicodemo AC, Araujo MR, Ruiz AS, Gales AC. In vitro susceptibility of Stenotrophomonas maltophilia isolates: comparison of disc diffusion, Etest and agar dilution methods. J Antimicrob Chemother. 2004;53:604–8.
Falagas ME, Valkimadi PE, Huang YT, Matthaiou DK, Hsueh PR. Therapeutic options for Stenotrophomonas maltophilia infections beyond co-trimoxazole: a systematic review. J Antimicrob Chemother. 2008;62:889–94.
Poulos CD, Matsumura SO, Willey BM, Low DE, McGeer A. In vitro activities of antimicrobial combinations against Stenotrophomonas (Xanthomonas) maltophilia. Antimicrob Agents Chemother. 1995;39:2220–3.
Liaw SJ, Teng LJ, Hsueh PR, Ho SW, Luh KT. In vitro activities of antimicrobial combinations against clinical isolates of Stenotrophomonas maltophilia. J Formos Med Assoc. 2002;101:495–501.
Acknowledgments
This study was supported by Hacettepe University Research Foundation (Project No: 060 D03 101 003).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Gülmez, D., Çakar, A., Şener, B. et al. Comparison of different antimicrobial susceptibility testing methods for Stenotrophomonas maltophilia and results of synergy testing. J Infect Chemother 16, 322–328 (2010). https://doi.org/10.1007/s10156-010-0068-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10156-010-0068-2