Abstract
Endobiotic bacteria colonize the tentacles of cnidaria. This paper provides first insight into the bacterial spectrum and its potential of pathogenic activities inside four cnidarian species. Sample material originating from Scottish waters comprises the jellyfish species Cyanea capillata and C. lamarckii, hydrozoa Tubularia indivisa and sea anemone Sagartia elegans. Mixed cultures of endobiotic bacteria, pure cultures selected on basis of haemolysis, but also lyophilized samples were prepared from tentacles and used for DGGE-profiling with subsequent phylogenetic analysis of 16S rDNA fragments. Bacteria were detected in each of the cnidarian species tested. Twenty-one bacterial species including four groups of closely related organisms were found in culture material. The species within these groups could not be differentiated from each other (one group of Pseudoalteromonas spp., two groups of Shewanella spp., one group of Vibrio spp.). Each of the hosts exhibits a specific endobacterial spectrum. Solely Cyanea lamarckii harboured Moritella viscosa. Only in Cyanea capillata, members of the Shewanella group #2 and the species Pseudoalteromonas arctica, Shewanella violacea, Sulfitobacter pontiacus and Arcobacter butzleri were detected. Hydrozoa Tubularia indivisa provided an amazingly wide spectrum of nine bacterial species. Exclusively, in the sea anemone Sagartia elegans, the bacterial species P. aliena was found. Overall eleven bacterial species detected were described recently as novel species. Four 16S rDNA fragments generated from lyophilized material displayed extremely low relationship to their next neighbours. These organisms are regarded as members of the endobiotic “terra incognita”. Since the origin of cnidarian toxins is unclear, the possible pathogenic activity of endobiotic bacteria has to be taken into account. Literature data show that their next neighbours display an interesting diversity of haemolytic, septicaemic and necrotic actions including the production of cytotoxins, tetrodotoxin and R-toxin. Findings of haemolysis tests support the literature data. The potential producers are Endozoicimonas elysicola, Moritella viscosa, Photobacterium profundum, P. aliena, P. tetraodonis, Shewanella waksmanii, Vibrio splendidus, V. aestuarius, Arcobacter butzleri.
Similar content being viewed by others
Introduction
Interactions between bacteria and invertebrates are common in marine environment (Deming and Colwell 1982; Paul et al. 1986; Cary et al. 1997; Burnett and McKenzie 1997; Althoff et al. 1998; Schuett et al. 2005; Wichels et al. 2006). Yet, the bacterial mode of association (symbiosis, commensalisms or parasitism) and particular physiological functions are often unidentified. This also applies to the scarcely investigated cnidaria from cold to moderate regions. Jellyfish represent a major zooplanktonic fraction in the North Sea and the Atlantic. Among them are dangerous stingers causing painful dermal injuries after contact with humans. Similarly, the sessile cnidarian sea anemones and hydrozoan species harbour complex toxin cocktails, mostly of unknown chemical structures and functions. Currently, these structures and activities of jellyfish species Cyanea capillata and C. lamarckii collected from the North Sea and the Atlantic are under investigation (Helmholz et al. 2007). However, the origin of cnidarian toxins is still open. A hypothesis favours the involvement of endobiotic bacteria as producers of cnidarian pathogenic activities. Reports of Palincsar et al. (1989) and Schuett et al. (2007) allowed first insight into the occurrence and morphology of organ-like bacterial aggregates in the intra-tentacular epidermis region of the sea anemone species Aiptasia pallida and Metridium senile.
This report provides an overview of cnidarian endobiotic bacteria from cold to moderate regions by phylogenetic analysis and allows information on their pathogenic potential.
Materials and methods
Samples and preparation
Individuals of Cyanea lamarckii, Cyanea capillata, Tubularia indivisa and Sagartia elegans were collected by divers during research cruises (June/July 2005–2008) from waters around the Orkney Islands. Tips of fresh tentacles were clipped off and subjected to washing procedures and CTAB treatment (Schuett et al. 2007) in order to remove potentially contaminating epibiotic bacteria. Light microscopic inspection showed no bacteria attached to the surface of fresh tentacle material. After removing seawater, one set of tentacle material was placed in three liquid medium modifications of Medium #621 (DSMZ, German Type Culture Collection; Staley 1968), shown in Table 1. Each test tube containing one tentacle tip was incubated under aerobic conditions for 2 months at 18°C. A second set containing the same sample material and medium ingredients has been incubated at microaerophillic conditions (90% N2 and 10% CO2). Additionally, cultures were tested for haemolytic activity according to Smibert and Krieg (1984) by using 5% (v/v) sheep blood.
Positive haemolytic strains were tested for their cytolytic activity by agar diffusion test using cell culture L929 (protocol provided by Lindl 2002).
Finally, washed tentacle material was lyophilized for further DNA extraction.
DNA extraction and PCR amplification of 16S rDNA fragments
One millilitre aliquots of aerobic and microaerophillic mixed cultures, respectively, picked colonies from blood agar plates were used for nucleic acid extraction according to Anderson and Mc Kay (1983). Primers 341fc (5′-cgc ccg ccg cgc ccc gcg ccc ggc ccg ccg ccc ccg ccc ccc tac ggg agg cag cag-3′, clamp region underlined) and 907rwob (5′-ccg tca att cct ttr agt tt-3′; Muyzer et al. 1995) were applied to PCR.
Reaction mixture (100 μl) contained 2 U of Taq DNA polymerase, 1× polymerase buffer and 1× Master Enhancer (Eppendorf), 100 μM each dNTP, 0.4 μM each primer and 1 μl DNA. After an initial denaturation step at 94°C for 3 min, 20 cycles (1 min each step) followed: denaturation 94°C, annealing temperature starting at 65°C and decreasing by 0.5°C per cycle, elongation 68°C. Another 12 cycles were added this time using 55°C as annealing temperature. A final incubation at 68°C for 6 min was followed by 18 cycles lowering the temperature for 1°C per minute in order to avoid the formation of heteroduplices. Negative controls were carried out omitting template DNA; Escherichia coli J53 served as positive control. DNA quantity was checked on agarose gels.
FastDNA Kit (Bio101) and DNeasy Blood & Tissue Kit (Qiagen) were combined for DNA extraction of lyophilized tentacles. Lysing Matrix tubes including a ¼″ Ceramic Sphere and Garnet Matrix (Bio101) were supplemented by 1 ml CLS-TC solution and 100 mg lyophilized material. The tube was placed in a Fast Prep Instrument (FP120, Bio101), processed for 30 s at speed 4 and incubated according to the manufacturer’s manual. After the first centrifugation step, the supernatant was transferred to a new tube and complemented by 20 μl proteinase K of the DNeasy Blood & Tissue Kit. The extraction was continued following Qiagen instructions. PCR was performed using Hot Master Taq polymerase (Eppendorf) and adapting the protocol (initial denaturation 1 min., annealing 30 s., elongation 65°C).
DGGE was performed using the DCode electrophoresis system (Biorad). Preparation of polyacrylamide gels (15–70% resp. 25–55% denaturant gradient) and electrophoresis parameters (100 V, 15 h) were performed according to Muyzer et al. (1995). Bands were stained with SYBR-Gold (Molecular Probes) and excised. DNA was extracted from gel material and dissolved in 10 μl distilled water (Sambrook et al. 1989).
Re-amplification of DNA fragments from DGGE bands
In difficult cases, a further purification of DNA fragments could be obtained by re-amplifying the extracted DNA using again the forward primer with clamp (341fc), checking the accurate position of the band and excising it after an additional DGGE run. The extracted DNA was re-amplified for sequencing by using the Taq DNA polymerase but this time applying the forward primer without clamp (341f) and omitting the enhancer. PCR products were purified by using the Qiaquick PCR Purification Kit (Qiagen) following the instructions of the manufacturer’s protocol and eluted with 60 μl distilled water.
DNA sequencing of PCR products and comparative sequence analysis
Sequencing was carried out by Qiagen Sequencing Services/Hilden, Germany. Sequences were aligned by using the Align IR program and the advanced BLAST search program of the National Center of Biotechnology Information (NCBI) website http://www.ncbi.nlm.nih.gov/Blast to find closely related sequences.
Results
Phylogenetic analysis of mixed cultures
Cultures were prepared at low concentrations of organic matter, applying aerobic and micro-aerophillic conditions. Different media were selected in order to widen the spectrum of culturable bacterial endobionts (Table 1). Bacterial cultures could be obtained from all four cnidarian species. Regarding DGGE patterns, the influence of different media compositions on the bacterial spectrum was negligible.
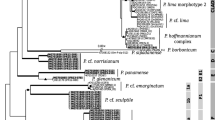
Seventy-nine fragments of the 16S rDNA deriving from cultures of four cnidarian species were successfully sequenced, aligned and phylogeneticly analyzed (consult Table 2). Most of the fragments displayed similarities between 98 and 100% to their next neighbours; exceptions were Endozoicimonas elysicola (97%), Arcobacter butzleri (97%) and Mesorhizobium tianshanense (89%). Seventeen bacterial species and four groups were found, the latter containing different closely related bacterial species which could not be differentiated from each other. The four groups were: (#1) Pseudoalteromonas tetraodonis/P. elyacovii/P. haloplanctis, (#2) Shewanella sairae/S. marinintestina, (#3) S.waksmanii/S. surugaensis/S. kaireiae and (#4) Vibrio splendidus/V. lentus/V. tasmaniensis/V. kanaloae. In these cases, the used fragment size was inappropriate to discriminate between the species forming these groups. Except for the Bacillus subtilis (Fermicutes), Ilyobacter psychrophilus (Fusobacterium) and Arcobacter butzleri (ε-proteobacterium) all other of the endobacteria detected were members of γ-proteobacteria. The most abundant endobionts comprised the groups #4 and #1 as well as Photobacterium profundum.
Remarkably, eleven bacterial species were described recently as novel organisms, some of them also experienced several taxonomic rearrangements. There were Cobetia marina (Arahal et al. 2002), Colwellia aestuarii (Jung et al. 2006), Endozoicimonas elysicola (Kurahashi and Yokoto 2006), Pseudoalteromonas aliena (Ivanova et al. 2004), Pseudoalteromonas marina (Nam et al. 2007), P. tetraodonis (Ivanova et al. 2001), Psychrobacter nivimaris (Heuchert et al. 2004), Shewanella livingstonensis (Bozal et al. 2002), S. sairae (Satomi et al. 2003), S. surugaensis (Miyazaki et al. 2006), S. violacea (Nogi et al. 1998). Their physiological traits are only partially known, yet.
Concluded, Tubularia indivisa provides the habitat for a wide spectrum of twelve bacterial species/groups, Cyanea capillata nine, Sagartia elegans eight and Cyanea lamarckii three.
Host-specific bacteria
All four of the cnidarian species tested harboured host-specific bacteria. Thirteen endobiotic species and groups occurred as single bacterial tenants, which are regarded as “specialists” for their individual hosts. Shewanella violacea, Sulfitobacter pontiacus, Arcobacter butzleri and members of group #2 were detected exclusively in C. capillata and Moritella viscosa only in C. lamarckii. Remarkable similarities between the jellyfish hosts could not be detected. The hydrozoa Tubularia indivisa sheltered the endobacteria Cobetia marina, Colwellia aestuarii, Endozoicimonas elysicola, Psychrobacter nivimaris, Shewanella livingstonensis, Vibrio aestuarianus, Bacillus subtilis and Ilyobacter psychrophilus; the sea anemone Sagartia elegans had Pseudoalteromonas aliena as host-specific tenant.
Phylogenetic analysis of lyophilized material
The phylogenetic analysis of six lyophilized samples comprising Cyanea capillata, C. lamarckii and Tubularia indivisa generated 10 bands. The band patterns displayed fewer bands than those of the cultures (Fig. 1). C. capillata fragments displaying Vibrio species group #4 were found in both samples (similarity 99%). Endozoicimonas elysicola (similarity 97%) was spotted in lyophilized but also in mixed culture material of Tubularia indivisa. This novel organism has also been detected previously in the sea anemone Metridium senile (Schuett et al. 2007). An exciting result was the discovery of endobacteria showing an extremely low relationship to their next neighbours. Because of high-sequence quality strange microorganisms detected in both of the Cyanea species and Tubularia indivisa should be interpreted as unknown new species. Among them were bacteria distant to Aquaspirillum itersonii (similarity 90%, found in Cyanea lamarckii), Francisella philomiragia (similarity 93%, both Cyanea species) and Spiroplasma syrphidicola, (89%, Tubularia indivisa). A definite phylogenetic allocation of these unambiguous sequence data to known bacteria is currently impossible.
Aggregates
Tubularia indivisa and Sagartia elegans contained organ-like bacterial aggregates in the tentacle’s epidermis. The aggregates are smaller than those of Aiptasia pallida (Palincsar et al. 1989) and Metridium senile (Schuett et al. 2007). Nevertheless, each of these aggregate-bearing cnidaria harbours a different bacterial spectrum. Cyanea species showed only free-floating endobiotic bacteria in their tentacles. A paper on aggregate visual nature and distribution by REM inspection is in preparation.
Pathogenic activities of bacterial endobionts
Nine bacterial species (out of 21) may possess pathogen activities and/or produce toxins (Table 3). This information has been taken from physiological literature data. It should be noted that the Pseudoalteromonas group #1 and the Vibrio group #4 contain members of pathogenic and non-pathogenic bacteria. All four cnidarian host organisms harbour potentially pathogen bacteria: C. lamarckii two species and C. capillata five, Tubularia indivisa and Sagartia elegans each five. Vibrio splendidus (the only pathogen species of group #4) has been detected in all of the cnidaria tested. Photobacterium profundum and Pseudoalteromonas tetraodonis (group #1) have been found in C. capillata, Tubularia indivisa and Sagartia elegans.
Some remarkable examples in detail: Hydrozoa Tubularia indivisa harbours the potentially pathogen Endozoicimonas elysicola which is suspected to play a role in amoebic fish disease (Bowman and Nowak 2004). Photobacterium profundum produces ToxR genes that mediate virulence expression (Bidle and Bartlett 2001; Ivanova et al. 2004). Pseudoalteromonas tetraodonis (group #1) is known to produce tetrodotoxin (Do et al. 1990; Ivanova et al. 2001). Metabolites of Vibrio aestuarianus may kill corals, oysters and turbot larvae. Extracellular toxic products of Vibrio aestuarianus (Labreuche et al. 2006; Garnier et al. 2007) may impair juvenile oysters. Members of Vibrio splendidus (group #4) can express septicaemic and necrotic activities (Sugumar et al. 1998; Thompson et al. 2003; Koren and Rosenberg 2006). Yet, it is uncertain whether anaerobic Ilyobacter psychrophilus, member of the genus Fusobacterium, generates severe clinical infection.
Anthozoa Sagartia elegans harbours Photobacterium profundum and Pseudoalteromonas aliena, which produces cytotoxins and causes haemolytic activities (Ivanova et al. 2004). Additionally, Sagartia elegans contains members of the groups #1 and #4 (activities see the previous paragraph).
Jellyfish Cyanea lamarckii provides the natural habitat for Moritella viscosa, which causes necrotic “winter ulcer” in salmons (Lunder et al. 2000; Heidarsdottir et al. 2008), additionally Arcobacter butzleri, a human pathogen species (Carbone et al. 2003; Gugliandolo et al. 2007), and finally members of group #4 (see the previous paragraph). The Lions mane jellyfish C. capillata carries the haemolytic species Shewanella waksmanii, a member of group #3 (Ivanova et al. 2003), and pathogenic endobionts of the already described Photobacterium profundum and the putatively pathogenic species of groups #1 and #4 (see the previous paragraph).
Haemolysis
Selection of mixed culture material based on haemolysis tests and subsequent phylogenetic analysis were applied to verify the pathogenic activities derived from literature data. The findings are in good correspondence with literature reports: haemolytic positives were detected in Cyanea capillata (Pseudoalteromonas group 1, 3 isolates; Vibrio group 4, 5 isolates), Tubularia indivisa (Vibrio group 4, 1 isolate), and in Sagartia elegans (Pseudoalteromonas aliena, 1 isolate; Pseudolateromonas group 1, 2 isolates; Vibrio group 4, 4 isolates). In order to determine cytolytic activity, the agar diffusion test was applied to haemolysis positive pure cultures. In case of Vibrio group #4 strain, derived from tentacle material of Cyanea capillata, the cells were completely disintegrated and decolorized within a 1-cm zone.
Discussion
The spectrum of culturable bacterial species is remarkable: some of them can be regarded as “all rounders”; others were host specific. An unambiguous identification on species level was excellent for seventeen different fragments (i.e., 80%). Unclear allocation of sequence data at species level led to the formation of groups # 1 to # 4. Some authors reported similar findings of different bacterial species (Ivanova et al. 2003 for Shewanella species; Marques et al. 2005; Venkateswaran et al. 1998 for Vibrio species; Mc Aucliffe et al. 2003 for Mycoplasma species). The application of larger 16S rDNA or 23S rDNA fragments might be more appropriate in future experiments. Physiological information concerning pathogenic activities was taken mainly from literature data and therefore insufficient as proof for pathogenic traits but also unsuitable in cases of unknown new species.
Eighty-one per cent of the species or groups found were host specific. This is a clear sign that bacterial contamination of the tentacle surface was avoided during experiments, which was confirmed by microscopic inspection subsequently to CTAB treatment.
Which approach reflects the actual presence of endobionts—phylogenetic data derived from cultures or lyophilized material? The strange, unidentified endobacteria may be regarded as the authentic tenants inhabiting the “exotic” tentacles whose physico-chemical conditions are still not discovered. Yet, PCR-processing of lyophilized material cannot detect low bacterial numbers. The major disadvantage of cultures is their medium dependence; the bacteria that grow may be of less endobiotic importance. In our experiments, the only bacterial species detected in both of the approaches was Endozoicimonas elysicola. To meet the actual growth requirements, refined culture techniques are needed.
The organ-like bacterial aggregates detected in Sagartia elgans and Tubularia indivisa contain hundreds of tightly packed, differently shaped endobacteria. Their different shapes suggest that these organisms may be host specific. The development and function of the organ-like aggregates are unclear. Do we presently observe here an evolutionary step during the formation of organ-like aggregate structures? Is that comparable to the developmental steps of prokaryotic mitochondria or chloroplasts in eukaryotes?
Data show that a considerable number of bacterial species living as tenants in cnidarian hosts may have pathogenic activity. The applied haemolysis and cell culture tests support the indirect approach using trait data from literature based on phylogenetic classification. Further testing of acute cell toxicity should be performed after calibrating bacterial protein concentration.
There is substantial knowledge of tetrodotoxin TTX, which is an extremely potent neurotoxin. Its chemical structure and activities at cellular level are well documented (Thuesen and Kogure 1989; Do et al. 1990; Ritchie et al. 2000). TTX-carrying animals are known from different marine, limnic and terrestrial habitats of warmer regions. Among them were the well-known puffer fish species (Tetraodontidae) but also starfish, toads, flatworms and even chaetognathes. Interestingly, the toxin is produced by endobiotic bacteria (Simidu et al. 1990; Yu et al. 2004). One of those endobionts is Pseudoalteromonas tetraodonis (Ivanova et al. 2001). Supposedly, these bacteria are also common partners of cnidarian species. In order to generate answers to the bacterial role in the production of cnidarian toxins, the investigation of their physiological traits is indispensable.
References
Althoff K, Schuett C, Krasko A, Steffen R, Batel R, Müller WEG (1998) Evidence for symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbour also putatively-toxic bacteria? Mar Biol 130:529–536
Anderson DG, Mc Kay LL (1983) Simple and rapid method for isolation of large plasmid DNA from lactic streptococci. Appl Environ Microbiol 46:549–552
Arahal DR, Castillo AM, Ludwig W, Schleifer KH, Ventosa A (2002) Proposal of Cobetia marina gen. nov., comb. nov. within the family Halomonadaceae to include the species Halomonas marina. Syst Appl Microbiol 25:207–211
Bidle KA, Bartlett DH (2001) RNA arbitrarily primed PCR survey of genes regulated by Tox R in the deep sea bacterium Photobacterium profundum strain SS9. J Bacteriol 183:1688–1693
Bowman JP, Nowak B (2004) Salmonid gill bacteria and their relationship to amoebic gill disease. J Fish Dis 27:483–492
Bozal N, Montes MJ, Tudela E, Jiménez F, Guinea J (2002) Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Arctic coastal waters. Int J Syst Evol Microbiol 52:195–205
Burnett WJ, McKenzie JD (1997) Subcuticular bacteria from the brittle star Ophiactis balli (Echinodermata, Ophiuroidea) represent a new lineage of extracellular marine symbionts in the α-subdivision of the class proteobacteria. Appl Environ Microbiol 63:1721–1724
Carbone M, Maugeri TL, Giannone M, Gugliandolo C, Midiri A, Fera MT (2003) Adherence of environmental Arcobacter butzleri and Vibrio spp. isolates to epithelial cells in vitro. Food Microbiol 20:611–616
Cary SC, Cottrell MT, Stein JL, Camacho F, Desbueres D (1997) Molecular identification and location of filamentous symbiotic bacteria associated with hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol 63:1124–1130
Deming J, Colwell RR (1982) Barophilic bacteria associated with digestive tracts of abyssal holothurians. Appl Environ Microbiol 44:1222–1230
Do HK, Kogure K, Simidu U (1990) Identification of deep sea sediment bacteria which produce tetrodotoxin. Appl Environ Microbiol 56:1162–1163
Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas J-L (2007) Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb Ecol 53:187–198
Gugliandolo C, Irrera GP, Lentini V, Maugeri TL (2007) Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the Straits of Messina (Italy). Mar Pollut Bull 56:600–606
Heidarsdottir KJ, Gravningen K, Benediktsdottir E (2008) Antigen profiles of the fish pathogen Moritella viscosa and protection in fish. J Appl Microbiol 104:944–951
Helmholz H, Ruhnau C, Schuett C, Prange A (2007) Comparative study on the cell toxicity and enzymatic activity of two northern scyphozoan species (Cyanea capillata (L) and Cyanea lamarckii (Pèron & Leslieur). Toxicon 50:53–64
Heuchert A, Glöckner FO, Amann R, Fischer U (2004) Psychrobacter nivimaris sp. nov., a heterotrophic bacterium attached to organic particles isolated from South Atlantic. Syst Appl Microbiol 27:399–406
Ivanova EP, Romanenko LA, Matté GR, Sysenko AM, Simidu U, Kita-Teukamoto K, Vysotskii MV, Frolova GM, Mikhailov V, Christen R, Colwell RR et al (2001) Retrieval of the species Alteromonas tetraodonis Simidu et al. 1990 as Pseudoalteromonas tetraodonis comb. nov. and amendation of description. Syst Evol Microbiol 51:1071–1078
Ivanova EP, Nedashkowskaya OI, Zhukova NV, Nicolau DV, Christian R, Mikhailov VV (2003) Shewanella waksmanii sp. nov., isolated from spuncula (Phascolosoma japonicum). Int J Syst Evol Microbiol 53:1471–1477. doi:10.1099/ijs.0.02630-0
Ivanova EP, Gorshkova NM, Zhukova NV, Lysenko AM, Zelepuga EA, Prokofeva NG, Mikhailov VV, Nicolau DV, Christen R (2004) Characterization of Pseudoalteromonas distincta-like sea-water isolates and description of Pseudoalteromonas aliena sp. Int J Syst Evol Microbiol 54:1431–1437
Jung SY, Oh TK, Yoon JH (2006) Colwellia aestuarii sp. nov., isolated from tidal flat sediment in Korea. Int J Syst Evol Microbiol 56:33–37
Koren O, Rosenberg E (2006) Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Micobiol 72:5254–5259
Kurahashi M, Yokoto A (2006) Endozoicimonas elysicola gen. nov., sp. nov., a γ-proteobacterium isolated from the sea slug Elysia ornata. Syst Appl Microbiol 30:202–206
Labreuche Y, Soudan P, Concalves M, Lambert C (2006) Effects of extracellular products from pathogenic Vibrio aestuarius strain 01/32 on lethality and cellular immune responses of the oyster Crassostrea gigas. In: Developmental and comparative immunology elsevier science, Oxford, pp 367–379
Lindl T (2002) Zell- und Gewebekultur. Spektrum Akademie Verlag Heidelberg, Berlin
Lunder T, Sørum H, Holstad G, Steigerwalt AG, Mowinckel P, Brenner DJ (2000) Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from atlantic salmon (Salmo salar) with winter ulcer. Int J Syst Evol Microbiol 50:427–450
Marques A, Dinh T, Iokeimidis C, Huys G, Swings J, Verstraete W, Dhont J, Sorgeloos P, Bossier P (2005) Effect of bacteria on Artemia franciscana cultured in different gnotobiotic environments. Appl Environ Microbiol 71:4307–4317
Mc Aucliffe L, Ellis RJ, Ayling RD, Nicolas RAJ (2003) Differentiation of mycoplasma species by 16S ribosomal DNA PCR and denaturant gradient gel electrophoresis fingerprinting. J Clin Microbiol 41:4844–4847
Miyazaki M, Nogi Y, Usami R, Horikoshi K (2006) Shewanella surugensis sp. nov., Shewanella kaireitica sp. nov. and Shewanella abyssi sp. nov. isolated from deep-sea sediments of Suruga Bay, Japan. Int J Syst Evol Microbiol 56:1607–1613
Muyzer G, Hottenträger S, Teske A, Wawer C (1995) Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communities. Mol Microb Ecol Manual 3.44, pp 1–22
Nam Y-D, Chang H-W, Park R, Kwon H-Y, Quan Z-X, Park Y-H, Lee J-S, Yoon J-H, Bae J-W (2007) Pseudoalteromonas marina comb. nov., a marine bacterium isolated from tidal flats of the Yello Sea, and reclassification of Pseudoalteromonas sagamiensis as Algicola sagamiensis comb. nov. Int J Syst Evol Microbiol 57:12–18
Nogi Y, Kato C, Horikoshi K (1998) Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch Microbiol 170:331–338
Palincsar EE, Jones WR, Palincsar JS, Glogowski MA, Mastro JL (1989) Bacterial aggregates within the epidermis of the sea anemone Aiptasia pallida. Biol Bull 177:130–140
Paul JH, De Flawn MF, Jeffrey WH (1986) Elevated levels of microbial activity in the corral surface microlayers. Mar Ecol Prog Ser 33:29–40
Ritchie KB, Nagelkerken I, James S, Smith GW (2000) Environmental microbiology: a tetrodotoxin-producing marine pathogen. Nature 404(6776):354
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Satomi M, Oikawa H, Yano Y (2003) Shewanella marinintestina sp. nov. Shewanella schlegelliana sp. nov. and Shewanella sairae sp. nov. eicosapentanoic-acid-producing marine bacteria isolated from sea animal intestines. Int J Syst Evol Microbiol 53:491–499
Schuett C, Doepke H, Groepler W, Wichels A (2005) Diversity of intratunical bacteria in the tunic matrix of the colonial ascidian Diplosoma migrans. Helgol Mar Res 59:136–140
Schuett C, Doepke H, Grathoff A, Gedde M (2007) Bacterial aggregates in the tentacles of the sea anemone Metridium senile. Helgol Mar Res 61:211–216
Simidu U, Kita-Tsukamoto T, Yasumoto T, Yotsu M (1990) Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int J Syst Bacteriol 40:331–336
Smibert RM, Krieg NR (1984) General characterization. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, p 432
Staley JT (1968) Prosthecomicrobium and Ancalomicrobium: new prosthcate freshwater bacteria. J Bacteriol 95:1921–1942
Sugumar G, Nakai T, Hirata Y, Matsubara D, Muroga K (1998) Vibrio splendidus var II as the causative agent of bacillary necrosis of Japanese oyster Crassostrea gigas larvae. Dis Aquat Organ 33:111–118
Thompson FL, Thompson CC, Swings J (2003) Vibrio tasmaniensis sp. nov. isolated from the Atlantic salmon (Salmo slar L.). Syst Appl Microbiol 26:65–69
Thuesen EV, Kogure K (1989) Bacterial production of tetrodotoxin in four species of Chaetognatha. Biol Bull 176:191–194
Venkateswaran K, Dohmoto N, Harayama S (1998) Cloning and nucleotide sequence of the gyr B Gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol 64:681–687
Wichels A, Wuertz A, Doepke H, Schuett C, Gerdts G (2006) Bacterial diversity in the breadcrump sponge Halichondria panicea (Pallas). FEMS Microbiol Ecol 56:102–118
Yu C-F, Yu PH-F, Chan P-L, Yan Q, Wong P-K (2004) Two novel species of tetrodotoxin- producing bacteria isolated from toxic marine puffer fishes. Toxicon 44:641–647
Acknowledgments
We are grateful to the BAH divers group who provided excellent fresh cnidarian sample material from difficult Atlantic locations. Special thanks go to Dr. Karl Herrmann (University Erlangen-Nürnberg) for his tireless microscopic hunt. Thanks are due to the crew of RV Heincke for their skilful engagement in difficult Scottish waters.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.-D. Franke.
Rights and permissions
About this article
Cite this article
Schuett, C., Doepke, H. Endobiotic bacteria and their pathogenic potential in cnidarian tentacles. Helgol Mar Res 64, 205–212 (2010). https://doi.org/10.1007/s10152-009-0179-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10152-009-0179-2