Abstract
Increasing rates of global trade and travel have the invariable consequence of an increase in the likelihood of nonindigenous species arrival, and some new arrivals are successful in establishing themselves. Quantifying the pattern of establishment of nonindigenous species across both spatial and temporal scales is paramount in early detection efforts, yet very difficult to accomplish. Previous work in epidemiology has proposed methods for assessing the space–time properties of emerging infectious diseases by quantifying the degree of space–time clustering between individual cases. I tested the applicability of one such method commonly used in epidemiology, the Knox test for space–time interaction, to analyze rare abundance data from an isolated, newly-establishing gypsy moth, Lymantria dispar, population in Minnesota, USA, and incorporated a bootstrap approach to quantify the space–time pattern in a random process that can be used to compare with results from empirical data. The use of the Knox test in assessing the establishment phase of biological invasions could potentially serve as an early warning system against new invaders, particularly for those with a known history of a repeated number of arrivals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The arrival and subsequent establishment of nonindigenous species in new habitats threatens native ecosystems, and their cascading direct and indirect effects are well documented (Parker et al. 1999; Mack et al. 2000; Pimentel et al. 2000; Mooney and Cleland 2001). Although one could argue that there is no shortage of invasion pathways through which nonindigenous species are continuously transported around the world (Liebhold et al. 2006; McCullough et al. 2006; Work et al. 2005), only a minority are believed to become established in new environments (Williamson and Fitter 1996; Ludsin and Wolfe 2001; Simberloff and Gibbons 2004), and only a proportion of these have inimical economical and environmental effects (Mack et al. 2000). Nevertheless, these adverse invasions, regardless of how rare, can have profound ramifications in native ecosystems.
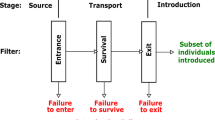
Distinguishing the potentially detrimental invasions from those that are more benign is not trivial, but also challenging is the recognition that an invader has indeed established itself. Following arrival, a new invader will either establish itself or not, and the probability of successful establishment can be affected by many factors, such as the size of the founder population, the susceptibility of the habitat to its invasion, the presence of competitors and regulators, environmental and demographic stochasticity, and Allee effects (Lockwood et al. 2007). Upon successful establishment, the next phase of the biological invasion process is spread, and the space–time properties associated with the spread of nonindigenous species have been extensively investigated (e.g., Okubo 1980; Andow et al. 1990; Lewis and Kareiva 1993; Shigesada and Kawasaki 1997). However, prior to modeling the spread of an invader is the determination that a new invasion is occurring. Thus, an analytical challenge is the quantification of the space–time properties associated with an emerging invasion and the determination that these properties are characteristic of a newly-established, reproducing population as opposed to one that arrived but failed to establish itself.
The ability to rapidly and quantitatively ascertain the existence and extent of an emerging biological invasion could more precisely dictate the appropriate management response, such as to deploy treatments aimed at eradication or to increase monitoring efforts. Upon their arrival, new invaders often exist at low abundance, which complicates survey and detection efforts. Moreover, the analysis of abundance data from initial infestations using conventional parametric approaches can be problematic because they are generally not robust enough to make any inference regarding establishment. Yet, when eradication of a new invader is a management goal, then a rapid response to the invasion is critical since the feasibility and costs of eradication are directly related to the degree of establishment (Rejmánek and Pitcairn 2002).
One approach in the analysis of rare or limited abundance data in biological invasions is to consider epidemiological methods, in which disease incidence can likewise be rare and limited in scale. One particular goal of invasion biologists and epidemiologists is not dissimilar; that is, to quickly detect the presence (or absence) of the space–time pattern associated with an establishing infestation. Such an early warning system may facilitate the implementation of aggressive management tactics prior to population increase and spread. Initial detection data often involve extremely low incidence levels, and are often limited temporally and spatially. Thus, in most cases, distributions of data are overwhelmingly dominated by zeros, highly skewed, and more often than not close to binary (i.e., presence/absence), all of which can supplement the inherent challenges of analyzing spatially and temporally autocorrelated data (Clifford et al. 1989). Many epidemiological methods, in contrast, were developed based on the incidence of a particular case of disease (i.e., presence), and therefore could have applicability to the analysis of rare data from biological invasions. In this paper, I used space–time data from a newly-establishing population of the gypsy moth, Lymantria dispar, a nonindigenous insect pest in North America, to investigate its space–time patterns using the fundamental basis of the Knox test for space–time interaction, a commonly used statistic in epidemiological investigations (Knox 1964a, b).
Materials and methods
Study system and data source
The gypsy moth, native to Eurasia, was introduced outside of Boston, Massachusetts, in 1869, and is now widely distributed over much of the eastern United States including portions west to Wisconsin and south to Virginia (Fig. 1) (Tobin et al. 2004). The gypsy moth is a defoliator of over 300 species of deciduous and coniferous host trees (Elkinton and Liebhold 1990). It is univoltine and overwintering eggs hatch in the spring. Adults generally emerge in mid- to late summer and, in the United States, females do not fly and generally oviposit within 1–2 m from the site of their emergence (Odell and Mastro 1980). Forms of dispersal include adult male flight, ballooning early instars, and the anthropogenic movement of gypsy moth life stages.
The area of the United States that is currently under quarantine for the gypsy moth, Lymantria dispar (gray area), 2005. The closed circle represents the city of Medford, where the gypsy moth was introduced in 1869. Its current distribution extends to Wisconsin (WI) to the west and Virginia (VA) to the south. Data from Minnesota (MN) were used in this analysis
Space–time data of a newly-establishing population was based upon count data of male gypsy moths measured from grids of spatially referenced pheromone-baited traps. Traps were deployed from 2000 to 2005 over an approximate 9,700 km2 area over portions of St Louis, Lake, and Cook counties in northeastern Minnesota, USA, along Lake Superior (Fig. 2), which is located at a considerable distance from the area currently under the United States gypsy moth quarantine (Fig. 1). Traps were set by the Minnesota Department of Agriculture as part of a United States Department of Agriculture national gypsy moth management program (cf. Tobin et al. 2004). Traps were generally placed approximately 2 km apart, though occasionally traps were placed 500–1,000 m apart when increased spatial resolution was required to more precisely determine the spatial extent of a possible gypsy moth infestation. The number of traps deployed in each year, and the corresponding frequency distribution of moths per trap, is presented in Table 1, which highlights the rarity in moth abundance but also a gradual increase in abundance that is perhaps common to many newly-establishing biological invaders.
Index of space–time interaction
One common statistical tool used in epidemiology to measure space–time clustering is the Knox test (Knox 1964a, b, 2002; Mantel 1967; Bailey and Gatrell 1995; Baker 1996, 2004; Kulldorff and Hjalmars 1999). This statistical tool is based on the notion that each pair of data, such as two cases of a disease, is separated by some measurable distance in both space and time. By using prespecified thresholds of spatial and temporal distance boundaries, it is possible to determine pairs of data that are concurrently “close” in space and time relative to pairs that are not (Fig. 3). A large number of pairs jointly close in space and time suggest space–time clustering, which furthermore often corresponds to the presence of an infectious process (Baker 1996). A summary of the use of Knox’s index in hypothesis testing is provided by Bailey and Gatrell (1995) and Baker (1996). In large data sets (Barbour and Eagleson 1986; Baker 2004), a null statistic can be developed by assuming that in the absence of space–time clustering (i.e., random distribution of events in space and time), then the number of observed pairs determined to be concurrently close in space and time (X) should approximate a Poisson distribution,
in which the mean of the distribution is equal to the product of the number of pairs that are close in space irrespective of time (S), and close in time irrespective of space (T), given n individual cases (Bailey and Gatrell 1995).
Application to biological invasions
Analogous to space–time clustering of an infectious disease could be the space–time patterns associated with the establishment of an invading organism. One critical limitation to Knox’s method is the requirement that space and time threshold boundaries, which are used to designate pairs of data concurrently close in space and time, are specified a priori (Bailey and Gatrell 1995; Baker 1996). In investigations of space–time processes, this is not trivial because it may not be known what is “close” in space and/or time. Rather than relying on prespecified threshold boundaries, I instead determined the number of pairs of data concurrently close in space and time across a range of spatial and temporal boundary thresholds (cf. Baker 1996). Thus, I varied the spatial threshold boundaries from 0.5 to 10 km in increments of 0.5 km (given the spatial resolution of deployed traps), used temporal threshold boundaries of 1, 2, or 3 years, and determined the number of pairs close in both space and time for each set of spatial and temporal threshold boundaries (i.e., 0.5 km and 1 year, 0.5 km and 2 year, ..., 10 km and 2 year, 10 km and 3 year).
Testing for space–time interaction
Baker (1996) developed an approach for a critical test statistic for use in hypothesis testing of space–time interaction, given that space–time data are unreplicable, and conceptually, I employed the same philosophy. In lieu of a prespecified statistical distribution, I used a bootstrapping approach to generate a theoretical distribution of space–time randomness with which empirical observations can be compared. Let N t be the total number of sampling locations (i.e., trap coordinates) in year t, and L t be the number of sampling locations that recorded a “presence” value, defined as a nonzero male moth capture from a trap. I then randomly selected L t spatial coordinate pairs (i.e., x and y coordinates) from N t . This was repeated for each year, in which t = 2000–2005. Pairs of spatial coordinates were selected with replacement. After selecting random locations for each year over the temporal ranges of the data, the number of pairs that were concurrently close in space and time were counted across the same spatial and temporal threshold boundaries used in the analysis of the empirical data. This constituted one iteration, and a distribution was based on 500 iterations of a random space–time series from which I estimated the median and 95% confidence intervals using the 2.5 and 97.5 percentiles of the distribution (Efron and Tibshirani 1993).
I also sought to derive a theoretical signature of the space–time pattern under an idealized space–time process. In this scenario, 0.5% of sampling locations from all traps deployed in Minnesota in 2000 were initially selected at random, and 0.5% was chosen because it was a conservative approximation of the initial percentage of traps in 2000 that recorded a nonzero moth capture (1.2%; Table 1). Then, using all trap locations from 2001, any location within 1 km of previously chosen locations was selected. Traps are generally deployed 2 km apart and, in practice, can be placed within a 500-m radius of the precise trap deployment coordinates. Thus, using 1 km as a threshold ensured that the same general locations were selected from year-to-year, and also captured new neighboring trap locations. This was continued through 2005 and constituted one iteration. This approach simulated population persistence in space through time and dispersal to nearby locations in the absence of stochasticity and extinction. The number of pairs were counted across the same spatial threshold boundaries used in prior analyses of the Minnesota data (0.5–10 km, in increments of 0.5 km), but, based on prior results, only a temporal threshold boundary of 1 year was used. For each new iteration, a new 0.5% selection of locations from 2000 was used, and a distribution was estimated based on 500 iterations. Although ecologically unrealistic, this scenario was specifically used as an example of one extreme space–time process, with the other being the signature obtained from a completely random process. Analyses and simulations were conducted in R Development Core Team (2005).
Results
The empirical data from Minnesota indicated a high degree of space–time clustering across a range of spatial and temporal threshold boundaries, and the patterns were significantly different from those values obtained from a distribution based upon random space–time locations (Fig. 4). Increasing the spatial and temporal threshold boundaries intuitively revealed that increasing these boundaries, even for a random space–time process, resulted in an increase in the number of pairs of data concurrently close in space and time. This result suggested that the space–time pattern observed from the empirical data in Minnesota was evidence of an established, reproducing gypsy moth population as opposed to one that was a result of repeated and random introductions.
Space–time pattern based upon empirical data from Minnesota (solid circles and line), with the corresponding pattern based upon a random process (solid line with dashed 95% CI) across a range of spatial (along the x-axis) and temporal (a, b, and c) boundary thresholds.1The number of pairs of data that are concurrently within the designated spatial and temporal threshold boundaries, divided the total number of observations (n = 933)
Although the empirical data indicated a space–time pattern that was significantly different from a random space–time process, it was also quite different from an idealized population simulated when assuming space–time persistence and in the absence of stochasticity and extinction (Fig. 5). This was somewhat expected because many biological invasions are influenced by the presence of stochasticity—demographic and environmental—and particularly by the role that Allee effects plays in newly founded colonies (Keitt et al. 2001; Liebhold and Bascompte 2003; Lockwood et al. 2005; Whitmire and Tobin 2006). Moreover, despite the bias in traps towards selecting males, which, because females are not sampled, may overestimate the range of reproducing populations, there is evidence supporting successful establishment in this particular region.
The empirical space–time pattern based upon empirical data from Minnesota (solid circles and line) compared with the corresponding patterns from both a random (solid line with dashed 95% CI) and idealized (solid line with open squares and dashed 95% CI) space–time process. Only a temporal threshold boundary of one year is shown.1The number of pairs of data that are concurrently within the designated spatial threshold boundaries and 1 year, divided by the total number of observations (n = 933)
The ecology and seasonality of the gypsy moth suggested that temporal and spatial threshold boundaries for analyzing the gypsy moth data could have been approximated and prespecified. For example, gypsy moth univoltinism would support defining a temporal threshold of 1 year. Similarly, movement in space is thought to be limited due to the flightless females, suggesting small spatial thresholds. However, an exploration of the space–time interaction over a range of values was a critical component to understanding the space–time interaction when applied to biological invasions. The fact that the empirical space–time pattern was consistently different, regardless of the spatial and temporal boundary threshold (Fig. 4), from the pattern obtained from a random process provides more evidence of an established gypsy moth population in this portion of Minnesota.
Discussion
The space–time pattern for an invasion process in which repeated introductions are spatially and temporally random should be distinctively different those in which successful establishment followed some introduction event. However, it is not always trivial to determine when an invader has successfully established itself in a new area, and in many cases, data upon which to base an assessment are limited. The potential of a nonindigenous species to invade new habitats can be considerably complex, and is often dependent on the details of the organism’s natural history (Crawley et al. 1986; Rejmánek and Richardson 1996; Goodwin et al. 1999; Kolar and Lodge 2001). Invading biological organisms thus may exhibit considerable variability in their respective ability to establish. For example, bark beetles (Coleoptera: Scolytidae) that depend upon mass attacking mechanisms to overwhelm host tree defenses (e.g., Raffa and Berryman 1983) may have a low invasion probability because of the density requirements of the founder population. In contrast, parthenogenic organisms could have a particularly high level of invasiveness because recognized causes of Allee effects, such as the difficulty in finding mates at low population densities, are inconsequential. Allee effects collectively refer to a decline in the growth rate of a population with a decline in its density, and causes include the inability to locate mates, inbreeding depression, and failure to satiate predators (Courchamp et al. 1999). Allee effects have been observed to play an important role in the establishment of isolated gypsy moth colonies (Liebhold and Bascompte 2003; Whitmire and Tobin 2006) and in its spread (Johnson et al. 2006; Tobin et al. 2007b). Knowledge of a species’ invasion potential, though critical, is not generally known, and sound management decisions could be optimized if an accurate assessment of establishment is made.
The gypsy moth invasion in North America provides a robust motivational data set upon which to determine if epidemiological approaches, such as the Knox index of space–time interaction, could be applied to biological invasions. Gypsy moth is one of the best-documented biological invasions in North America (Tobin et al. 2007a). Newly-establishing populations are continuously monitored through the extensive deployment of grids of pheromone-baited traps ahead of the endemic area (Tobin et al. 2004), which provides a data source that can be used to assess a space–time pattern during the initial invasion into a new area, and thus potentially reflects the process that is common to other biological invasions. Future work could use this motivational data set to explore the applicability of other epidemiological methods in analyzing the establishment of biological invaders, such as the concept of hazard modeling (Lawson and Denison 2002).
Epidemiologists have long been interested in the spatial and temporal properties of an emerging infectious disease, and map-making approaches to facilitate this assessment date at least as far back to the 1854 cholera epidemic in London (Brody et al. 2000). Statistical approaches in quantifying spatial pattern formation of disease dynamics are theoretically and empirically robust (Knox 1964a, b; Mantel 1967; Marshall 1991; Baker 1996, 2004), and are based upon the strong tendency for cases of infectious diseases to be clustered over spatial and temporal scales (e.g., Grenfell et al. 2001). Biological invasions, particularly in newly-establishing populations, can also be inherently clustered, and methods developed within the context of epidemiological investigations could also be applicable in the study of invasions, particularly in cases where a new invader is continuously being detected. In such cases, the question remains if repeated detections are simply the result of repeated, and random, introductions, or if establishment was successful following introduction. Because the management decision could be completely different depending on whether or not establishment was successful, a quantitative and statistically robust approach to assessing establishment, even with limited data, could be valuable.
The concept of an early warning system to rapidly detect an emerging threat, such as an increase in disease incidence, has been previously suggested as a basis for the development of a rapid response to the threat (Mostashari et al. 2003), and many invasion biologists have a similar goal. However, since resources to tackle invasions are more often than not binding, resource prioritization is extremely critical in management efforts and particularly for those that attempt eradication. Environmental and demographic stochasticity, coupled with Allee effects, can play important roles in the establishment of a new invader and, hence, not all invasions are successful. Moreover, the time from initial arrival to establishment can vary from species to species. Using epidemiological methods, such as the one I explored in this paper, to quantify the spatial and temporal patterns in biological invasions could accomplish the goals of identifying a newly establishing invader, which may determine if post-detection surveys are necessary, and assessing the relative degree of establishment, which facilitates the development of an appropriate and feasible management response.
References
Andow DA, Kareiva PM, Levin SA, Okubo A (1990) Spread of invading organisms. Landsc Ecol 4:177–188
Bailey TC, Gatrell AC (1995) Interactive spatial data analysis. Longman, Essex
Baker RD (1996) Testing for space–time clusters of unknown size. J Appl Stat 23:543–554
Baker RD (2004) A modified Knox test of space–time clustering. J Appl Stat 31:457–463
Barbour AD, Eagleson GK (1986) Tests for space–time clustering. Lect Notes Math 1212:42–51
Brody H, Rip MR, Vinten-Johansen P, Paneth N, Rachman S (2000) Map-making and myth-making in broad street: the London Cholera epidemic, 1854. Lancet 356:64–68
Clifford P, Richardson S, Hémon D (1989) Assessing the significance of the correlation between two spatial processes. Biometrics 45:123–134
Courchamp F, Clutton-Brock T, Grenfell B (1999) Inverse density dependence and the Allee effect. Trends Ecol Evol 14:405–410
Crawley MJ, Kornberg H, Lawton JH, Usher MB, Southwood R, O’Connor RJ, Gibbs A (1986) The population biology of invaders. Philos Trans R Soc Lond B 314:711–731
Efron B, Tibshirani RJ (1993) An introduction to the bootstrap. Chapman and Hall, London
Elkinton JS, Liebhold AM (1990) Population dynamics of gypsy moth in North America. Annu Rev Entomol 35:571–596
Goodwin BJ, McAllister AJ, Fahrig L (1999) Predicting invasiveness of plant species based on biological information. Conserv Biol 13:422–426
Grenfell BT, Bjørnstad ON, Kappey J (2001) Travelling waves and spatial hierarchies in measles epidemics. Nature 414:716–723
Johnson DM, Liebhold AM, Tobin PC, Bjørnstad ON (2006) Allee effects and pulsed invasion by the gypsy moth. Nature 444:361–363
Keitt TH, Lewis MA, Holt RD (2001) Allee effects, invasion pinning, and species borders. Am Nat 157:203–216
Knox EG (1964a) The detection of space–time interactions. Appl Stat 13:25–30
Knox EG (1964b) Epidemiology of childhood leukaemia in Northumberland and Durham. Br J Prev Soc Med 18:17–24
Knox EG (2002) An epidemic pattern of murder. J Public Health Med 24:34–37
Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Trends Ecol Evol 16:199–204
Kulldorff M, Hjalmars U (1999) The Knox method and other tests for space–time interaction. Biometrics 55:544–552
Lawson AB, Denison DGT (2002) Spatial cluster modelling. Chapman and Hall, Boca Raton
Lewis MA, Kareiva P (1993) Allee dynamics and the spread of invading organisms. Theor Popul Biol 43:141–158
Liebhold AM, Bascompte J (2003) The Allee effect, stochastic dynamics and the eradication of alien species. Ecol Lett 6:133–140
Liebhold AM, Work TT, McCullough DG, Cavey JF (2006) Airline baggage as a pathway for alien insect species entering the United States. Am Entomol 52:48–54
Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20:223–228
Lockwood JL, Hoopes MF, Marchetti MP (2007) Invasion ecology. Blackwell, Malden
Ludsin SA, Wolfe AD (2001) Biological invasion theory: Darwin’s contributions from The Origin of Species. Bioscience 51:780–789
Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA (2000) Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl 10:689–710
Mantel N (1967) Detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marshall RJ (1991) A review of methods for the statistical analysis of spatial patterns of disease. J R Stat Soc A 154:421–441
McCullough DG, Work TT, Cavey JF, Liebhold AM, Marshall D (2006) Interceptions of nonindigenous plant pests at US ports of entry and border crossings over a 17-year period. Biol Invasions 8:611–630
Mooney HA, Cleland EE (2001) The evolutionary impact of invasive species. Proc Natl Acad Sci USA 98:5446–5451
Mostashari F, Kulldorff M, Hartman JJ, Miller JR, Kulasekera V (2003) Dead bird clusters as an early warning system for West Nile virus activity. Emerg Infect Dis 9:641–646
Odell TM, Mastro VC (1980) Crepuscular activity of gypsy moth adults (Lymantria dispar). Environ Entomol 9:613–617
Okubo A (1980) Diffusion and ecological problems: mathematical models. Springer, Berlin Heidelberg New York
Parker IM, Simberloff D, Lonsdale WM, Goodell K, Wonham M, Kareiva PM, Williamson MH, Von Holle B, Moyle PB, Byers JE, Goldwasser L (1999) Impact: toward a framework for understanding the ecological effects of invaders. Biol Invasions 1:3–19
Pimentel D, Lach L, Zuniga R, Morrison D (2000) Environmental and economic costs of nonindigenous species in the United States. Bioscience 50:53–65
R Development Core Team (2005) http://www.r-project.org
Raffa KF, Berryman AA (1983) The role of host plant resistance in the colonization behavior and ecology of bark beetles (Coleoptera: Scolytidae). Ecol Monogr 53:27–49
Rejmánek M, Pitcairn J (2002) When is eradication of exotic pest plants a realistic goal? In: Veitch D, Clout M (eds) Turning the tide: the eradication of invasive species, IUCN SSC invasive species specialist group, pp 249–253
Rejmánek M, Richardson DM (1996) What attributes make some plant species more invasive? Ecology 77:1655–1661
Shigesada N, Kawasaki K (1997) Biological invasions: theory and practice. Oxford University Press, Oxford
Simberloff D, Gibbons L (2004) Now you see them, now. you don’t! Biol Invasions 6:161–172
Tobin PC, Sharov AA, Leonard DS, Roberts EA, Liebhold AM (2004) Management of the gypsy moth through a decision algorithm under the Slow-the-Spread project. Am Entomol 50:200–209
Tobin PC, Liebhold AM, Roberts EA (2007a) Comparison of methods for estimating the spread of a nonindigenous species. J Biogeogr 34:305–312
Tobin PC, Whitmire SL, Johnson DM, Bjørnstad ON, Liebhold AM (2007b) Invasion speed is affected by geographic variation in the strength of Allee effects. Ecol Lett 10:36–43
Whitmire SL, Tobin PC (2006) Persistence of invading gypsy moth colonies in the United States. Oecologia 147:230–237
Williamson M, Fitter A (1996) The varying success of invaders. Ecology 77:1661–1666
Work TT, McCullough DG, Cavey JF, Komsa R (2005) Arrival rate of nonindigenous insect species in the United States through foreign trade. Biol Invasions 7:323–332
Acknowledgments
I thank Laura Blackburn and Gino Luzader (USDA Forest Service); E. Anderson Roberts (Virginia Tech); and Kimberly Thielen Cremers (Minnesota Department of Agriculture) for their assistance in data acquisition and compilation. I also thank Robert C. Venette and R. Talbot Trotter (USDA Forest Service), and Abigail Walter (University of Minnesota) for providing thoughtful reviews of this paper prior to submission.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tobin, P.C. Space–time patterns during the establishment of a nonindigenous species. Popul Ecol 49, 257–263 (2007). https://doi.org/10.1007/s10144-007-0043-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-007-0043-7