Abstract
Low temperature is a major factor limiting rice growth and yield, and seedling is one of the developmental stages at which sensitivity to chilling stress is higher. Tolerance to chilling is a complex quantitative trait, so one of the most effective approaches to identify genes and pathways involved is to compare the stress-induced expression changes between tolerant and sensitive genotypes. Phenotypic responses to chilling of 13 Japonica cultivars were evaluated, and Thaibonnet and Volano were selected as sensitive and tolerant genotypes, respectively. To thoroughly profile the short-term response of the two cultivars to chilling, RNA-Seq was performed on Thaibonnet and Volano seedlings after 0 (not stressed), 2, and 10 h at 10 °C. Differential expression analysis revealed that the ICE-DREB1/CBF pathway plays a primary role in chilling tolerance, mainly due to some important transcription factors involved (some of which had never been reported before). Moreover, the expression trends of some genes that were radically different between Thaibonnet and Volano (i.e., calcium-dependent protein kinases OsCDPK21 and OsCDPK23, cytochrome P450 monooxygenase CYP76M8, etc.) suggest their involvement in low temperature tolerance too. Density of differentially expressed genes along rice genome was determined and linked to the position of known QTLs: remarkable co-locations were reported, delivering an overview of genomic regions determinant for low temperature response at seedling stage. Our study contributes to a better understanding of the molecular mechanisms underlying rice response to chilling and provides a solid background for development of low temperature-tolerant germplasm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the most important crops cultivated in both tropical and temperate regions, since its grain represents a staple food for more than half of the world population (FAO 2015). With 234,134 ha of cultivated area in 2016 (Ente Nazionale Risi 2017), Italy is the first rice producer in Europe. Rice is highly sensitive to low temperature (Andaya and Tai 2006), and chilling is a major factor limiting its productivity (Zhao et al. 2013). The world’s population is going to largely grow in the next decades, thus stabilizing rice production by increasing tolerance to low temperatures would be crucial.
Seedling is one of the rice developmental stages at which thermal sensitivity is higher, especially in temperate regions where low temperatures occurring after rice sowing may drastically affect crop productivity (Andaya and Mackill 2003; Ji et al. 2008). Chilling stress affects chlorophyll content, interferes with photosynthesis (Kanneganti and Gupta 2008; Kim et al. 2009), and causes accumulation of reactive oxygen species (ROS) and malondialdehyde (MDA) that can impair metabolism activity via cellular oxidative damage (Nakashima et al. 2007; Xie et al. 2009). Rice can also tolerate cold stress: for example, cold-treated rice plants accumulate the amino acid proline and antioxidant species that are able to stabilize protein synthesis and scavenge ROS (Sato et al. 2011). Accessions belonging to the Japonica group usually show higher levels of cold tolerance compared to Indica ones (Mackill and Lei 1997; Pan et al. 2013). A better understanding of plants molecular response to chilling stress is thus essential to breed tolerant rice cultivars through genetic improvement.
From a genetic point of view, more than 250 quantitative trait loci (QTL) controlling cold tolerance were identified on all 12 rice chromosomes at different growth and developmental stages (Mao et al. 2015; Xiao et al. 2015; Yang et al. 2015; Zhu et al. 2015), suggesting that cold tolerance in rice is a complex quantitative trait (Maruyama et al. 2014), and appears more complicated in comparison to what observed for other Poaceae species (Pecchioni et al. 2014). Among these QTLs, several have been fine mapped, including some QTLs related to low temperature tolerance at seedling stage (Andaya and Tai 2006, 2007; Koseki et al. 2010; Kim et al. 2014; Xiao et al. 2015). One of the most interesting loci—named COLD1—confers chilling tolerance in Japonica rice seedlings and encodes a regulator that interacts with the G-protein α subunit to activate the Ca2+ channel for sensing low temperature and accelerate G-protein GTPase activity (Ma et al. 2015). Recently, Wang et al. (2016) performed a genome-wide association study (GWAS) for cold tolerance at seedling stage using 295 rice cultivars of different subspecies and found 67 QTLs on 11 chromosomes (46 of which were located in genomic regions lacking previously known QTLs). Most of the genomic regions determining rice response to low temperature are reported in the Gramene QTL database (http://archive.gramene.org/qtl/), showing loci spanning all chromosomes putatively linked to the trait.
From a molecular and physiological point of view, there are three known main pathways through which rice can react to low temperature (Zhang et al. 2014): an ICE-DREB1/CBF pathway, an ABA-dependent pathway and a MAPK cascade. The DREB1/CBF (dehydration responsive element binding/cold-binding factor) cascade is the most characterized one and involves the upstream regulator ICE (Inducer of CBF Expression) and the DREB1/CBF transcription factors that modulate the downstream COR (Cold-Regulated genes) effectors (Zhou et al. 2011). DREB1/CBF transcription factors specifically interact with the cis-acting C-repeat/dehydration-responsive (CRT/DRE) elements and control the expression of whole sets of COR genes called the “CBF regulon” (Chinnusamy et al. 2007; Zhou et al. 2011; Pan et al. 2013; Pecchioni et al. 2014). In rice, genes OsDREB1A and OsDREB1B are early induced upon chilling stress, while OsDREB1C was reported to be not modulated (Dubouzet et al. 2003). Another DREB1/CBF, OsDREB1D, is involved in the ABA-dependent pathway activated in response to cold (Haake et al. 2002), thus representing a crossroad of signaling transduction cascades. Abscisic acid affects the expression of ABA-responsive genes via cis-acting ABA-response elements (ABRE) and the ABRE-binding bZIP transcription factors (ABF) (Hossain et al. 2010). OsNAC transcription factors transduce the ABA signal and regulate the expression of genes containing the NAC recognition sequence (NACRS) increasing cold tolerance in rice (Nakashima et al. 2007, 2012; Song et al. 2011). The mitogen-activated protein kinase (MAPK) cascade is the third cold response pathway identified in plants (Boudsocq and Laurie 2005): in cold-treated rice cells, accumulation of reactive oxygen species (ROS) triggers a downstream cascade involving MAPK genes (OsMKK6-OsMPK3) that affects the expression of target COR genes (Xie et al. 2009).
Advances in large scale RNA-Seq provided highly efficient and low cost methods to analyze whole transcriptomes, allowing a better understanding of chilling response genetic control: in recent years, several comparisons between the transcriptomes of tolerant and sensitive rice plants in stressed and non-stressed conditions have been made. Da Maia et al. (2016) used this technique with plants at germination stage, comparing transcriptomes of Indica and Japonica genotypes. Shen et al. (2014) used seedlings of four contrasting genotypes: Indica 93-11 cultivar as cold sensitive, and three cold-tolerant accessions (Dongxiang and Chaling wild rices, and an F2 line derived from a cross between Dongxiang and 93-11). Wang et al. (2017) studied differentially expressed genes (DEGs) in a tolerant chromosome segment substitution line and in its sensitive recurrent parent 93-11 under chilling stress. Results of these comparisons between tolerant and sensitive genotypes in control and stress conditions expanded our understanding of the complex mechanisms involved in chilling tolerance in rice but, to the authors’ knowledge, no studies comparing the transcriptome response of two contrasting cultivated Japonica genotypes have been reported so far. Moreover, all available previous studies compared transcriptomes of contrasting genotypes under standard and low temperatures conditions only (Zhang et al. 2012; Ma et al. 2015; Xiao et al. 2015; da Maia et al. 2016),without providing any information on gene expression changes at different time-points after chilling exposure.
The aims of this study are to profile the short-term transcriptional response of rice to chilling at different time-points and to identify genes that are putatively responsible for cold stress-tolerance. Twelve rice varieties cultivated in Italy, together with reference cultivar Nipponbare, were exposed to chilling stress at the seedling stage, monitoring phenotypic traits related to tolerance/sensitivity. On these bases, two contrasting genotypes were selected and used for further experiments. The short-term molecular responses of the two genotypes to low temperatures at two different time-points (referred to as “early” and “late”) between those two cultivars were analyzed by RNA-Seq. The results of our studies may contribute to elucidate the molecular mechanisms involved in rice response to chilling and to identify novel genes putatively involved in low temperature tolerance.
Materials and methods
Plant material and phenotypic evaluation
Thirteen rice (Oryza sativa ssp. Japonica) cultivars namely Baldo, Arborio, Augusto, Balilla. Carnaroli, Eurosis, Gange, Loto, S. Andrea, Thaibonnet, Vialone Nano, Volano, and the reference cultivar Nipponbare, were tested in controlled conditions to observe the effects of chilling stress and to identify two contrasting genotypes for the following transcription profiling studies. Seeds were kindly provided by Dr. Giampiero Valé of the Council for Agricultural Research and Economics, Research Centre for Cereal and Industrial Crops (CREA-CI), Vercelli, Italy. Three different experiments were carried out in growth chamber: the first one was designed to test the effects of several stress durations, while second and third ones were performed (according to first experiment results) in order to select a tolerant and a sensitive genotype among the 13 cultivars.
Experiment 1
Rice seeds were germinated on filter paper soaked in sterile water at 28 °C in the dark for 4 days. Five seedlings for each of the 13 genotypes were transplanted in pots filled with water-saturated soil and moved for 4 days to a growth chamber (Binder KBW 720, Tuttlingen, Germany) with a 14-h photoperiod under an irradiance of 180 μmol m−2 s−1 (white fluorescent tubes Fluora 18 W/77, Osram, Munich, Germany), day/night temperatures of 28/20 °C, and a relative humidity of 60%. Soil was then covered with a 2 cm water layer and plants—rotated around the chamber daily to minimize within-chamber effects—were grown at the same (control) conditions up to 2-leaves stage. To observe the effects of stress at different durations, chilling (10 °C) was applied for 0, 2, 7, and 14 days at 28/10 °C (day/night temperatures) under a 14-h day/10-h night cycle. Three biological replicates (five plants each) were carried out, and the whole experiment was repeated twice. Electrolyte leakage test was used to assess cell membrane stability according to Caffagni et al. (2014): briefly, freshly cut leaf discs (ca. 0.5 cm diameter) were placed in vials containing 25 cm3 of deionized water and stirred at 25 °C for 2 h 30 min. The conductivities of the solution (C1) and of deionized water (Cw) were determined with a digital conductance meter (GLP 31, Crison Instruments, Barcelona, Spain); samples were then autoclaved at 120 °C for 15 min, and conductivity (C2) was measured again to determine electrolyte leakage (%) according to the formula: EL(%) = (C1-Cw)/(C2-Cw) × 100.
Experiment 2
Adopting the same growing conditions used in Experiment 1, three traits were measured after 7 or 14 days of chilling stress. Visual Score: plant health after cold stress was visually scored according to the IRRI-CTol guidelines (IRRI 2002); Leaf Area: plants were cut 2 cm from soil level and total leaf area was estimated with software GIMP 2.6 from images captured using a standard flatbed scanner at 300 dpi resolution; Electrolyte Leakage: measured as described in Experiment 1.
Experiment 3
The 13 genotypes were also characterized in an independent experiment in terms of shoot elongation under low-temperature stress following a modification of the growth protocol adopted by Niroula et al. (2012). Briefly, rice seeds were de-hulled, sterilized with 5% (v/v) hypochlorite and washed several times with sterile water. Seeds were maintained in 1%-agar plate ½ MS medium in the dark at 30 °C for 3 days. Seedlings were then grown in a growth cabinet under a 14-h-light photoperiod, PAR 50 μmol m−2 s−1 provided by fluorescent lamps under two different temperature regimes: (i) 28/28 °C day/night; (ii) 28/10 °C day/night. Shoot length measures were scored after 7 and 14 days of chilling stress.
All phenotypic data obtained from the described experiments were analyzed through GenStat 17th Edition software (Payne 2014). Linear mixed models and estimation of variance components using the method of residual maximum likelihood (REML) were applied. For each analysis, Levene’s test was carried out to assess the assumption of equality of variances of standardized residuals. Approximate least significant differences (LSD p value < 0.05) of REML means were then used for multiple comparisons.
RNA extraction and quantitative PCR
A preliminary real-time quantitative PCR (RT-qPCR) experiment was carried out to select the best two time-points for profiling short-term transcriptome response to chilling stress. Ten-day-old seedlings of Volano and Thaibonnet (the two cultivars chosen as tolerant and sensitive to chilling stress, respectively) were grown under a 14-h day/10-h night cycle under control (28/28 °C) and stress (28/10 °C). Samples were collected 0, 12, 16, 24, 36, 40, and 48 h after the beginning of the experiment, immediately frozen in liquid nitrogen and kept at − 80 °C until RNA isolation. Total RNA was extracted from shoots using Spectrum™ Plant Total RNA Kit (Sigma Aldrich, USA) and DNA was removed using the Turbo DNA-free™ Kit (Ambion by Life Technology, USA). DNAase-treated RNA was reverse-transcribed using the iScript™ cDNA Synthesis Kit (Biorad, USA), according to the manufacturer’s instructions.
OsDREB1A (Os09g0522200, The Rice Annotation Project Database, 2017-08-04) is well-known to be upregulated shortly after the initiation of chilling treatment (Mao and Chen 2012). In particular, an induction of OsDREB1A transcription is evident after less than 1 h of exposure to low temperatures (Dubouzet et al. 2003). OsMYB4 (Os01g0695900, The Rice Annotation Project Database, 2017-08-04) is reported to be induced later, between 6 and 12 h after the initiation of chilling treatment (Yun et al. 2010). On the basis of these evidences, we assumed OsDREB1A and OsMYB4 genes to act as models for “early” and “late” transcriptional short-term response to chilling stress, respectively. Their relative expression levels were measured via RT-qPCR in our cDNAs and used to identify two critical time-points, when “early” and “late” transcriptional differences between sensitive and resistant genotypes are more evident.
Primers for RT-qPCR were designed with Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012) using the following conditions: primer size 18–30 bp, primers Tm 58–62 °C, product Tm 70–90 °C, primer GC% 40–60%, product size 80–150 bp. NetPrimer software (Premier Biosoft, USA) was used to control hairpins, self dimers and cross dimers ΔG. Real time PCR was performed using the iTaq Universal SYBR Green Supermix (Biorad, USA) with primers specifically designed for OsDREB1A and OsMYB4 genes and Actin-1 as internal control (Online Resource 1). The relative expression levels of each gene were quantified as described in the ABI PRISM 7000 Sequence Detection System (User Bulletin No 2, Applied Biosystems, USA). RT-qPCR reactions were performed in duplicate.
RNA sequencing
Thaibonnet and Volano seedlings were grown adopting the same conditions described in the previous paragraph. Since the aim for RNA-Seq experiment was to observe the transcriptional changes in seedlings caused by stress, the control point was set right before the onset of the chilling treatment (i.e., 14 h after the beginning of the experiment). Total RNAs were extracted from Thaibonnet and Volano seedlings leaves at control time-point and after 2 and 10 h (i.e., 14, 16, and 24 h from the beginning of the time course experiment) using the method described above. Three biological replicates of RNA were extracted and eluted in TE buffer for each genotype and time point for a total of 18 RNA samples. Total RNAs concentration was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, USA) and RNAs integrity was assessed with Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Samples returning a RIN value > 8.0 were considered acceptable for sequencing. A total of 500 ng for each of the 18 high-quality RNA samples were sent in dry ice to Beckman Coulter Genomics, Inc. (Danvers, USA) for libraries construction, multiplexing and paired-ends sequencing with Illumina HiSeq (2 × 75 bps) using three chip lanes. After assessing their quality with FastQC (Andrews 2010), RNA-Seq reads have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5941.
Differential expression analysis
Trimmomatic (Bolger et al. 2014) was used to filter out adaptors sequences and low-quality bases, then filtered RNA reads were mapped to Oryza sativa ssp. Japonica (Nipponbare IRGSP-1.0) and Oryza sativa ssp. Indica cultivar 93–11 (Yu et al. 2002) genome assemblies with Bowtie2/TopHat2 aligner (Kim et al. 2013). Read counts were generated from alignment files with HTSeq software (Anders et al. 2015) in “intersection-nonempty” mode.
Differential expression analysis was carried out with EdgeR version 3.16.5 (Robinson et al. 2010) on the 37,830 rice genes (non-coding RNA genes were excluded). EdgeR was used to (i) filter out the not expressed or poorly expressed genes (we considered as “active” the genes with counts per million bases > 1 in at least 2 libraries), (ii) normalize the RNA libraries, and (iii) do the differential expression analysis with likelihood ratio test of 2- and 10-h treatments comparing to control samples for both Volano and Thaibonnet. False Discovery Rate (FDR) < 0.05 and log|FC| > 1.00 were considered as the conditions to state the genes as differentially expressed. Functional annotations of the rice transcripts were taken from RAP-DB website (http://rapdb.dna.affrc.go.jp/).
GO enrichment and MapMan analysis
GO enrichment analyses were conducted with GOseq Bioconductor package release 1.26.0 (Young et al. 2010). GOseq software is specifically designed to bias the RNA-Seq data by transcripts length, so median length of the transcripts for each gene were calculated with GenomicFeatures Bioconductor package version 1.26.2 (Lawrence et al. 2013) using Ensembl .gtf file as input (Ensembl plants; Nipponbare; Oryza_sativa.IRGSP-1.0.34). A .tsv file including the GO annotation of all the rice genes was retrieved from BioMart EnsemblPlants (Oryza sativa ssp. Japonica genes IRGSP-1.0). GO enrichment analysis was based on the Wallenius approximation, and only GO categories with p value < 0.05 were considered as enriched. MapMan 3.5.1 software (Usadel et al. 2009) was also used as a tool for pathway-based analysis to further characterize the genes that were upregulated earlier in Volano than in Thaibonnet.
Data availability
The RNA-Seq datasets generated and analyzed during the current study are available in the ArrayExpress database at EMBL-EBI under accession number E-MTAB-5941. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5941
Results
Cultivar phenotyping
Cold stress (10 °C during nighttime) was effective on the 13 tested rice cultivars, in terms of EL%, starting from the second day of treatment, but differences among varieties became significant at 7 and, even more, at 14 days (Online Resource 2). Electrolyte leakage, visual score, leaf area of seedlings and shoot length were then measured before chilling and after 7 and 14 days of overnight 10 °C treatment. The measured parameters showed different responses of rice cultivars to chilling stress (Table 1): according to statistical tests, Thaibonnet showed the highest sensitivity to chilling, while both Arborio and Volano resulted the most resistant genotypes. Therefore, Thaibonnet was chosen as sensitive cultivar for the gene expression profiling experiment and, after an overall evaluation, Volano was selected as tolerant (e.g., it is a more widely diffused cultivar in Italy than Arborio, 18,121.34 cultivated hectares vs. 919.6, according to 2016 statistics by Ente Nazionale Risi).
Stress time-points choice for RNA-Seq analysis
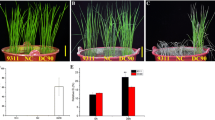
According to literature, we assumed OsDREB1A (Os09g0522200) and OsMYB4 (Os01g0695900) as representative of genes whose short-term transcriptional response to chilling is “early” and “late,” respectively. Yun et al. (2010) reported a strong increasing in OsMYB4 gene activity after 6–12 h at 10 °C, while Dubouzet et al. (2003) observed an induction of OsDREB1A gene transcription within 40 min after exposure to low temperatures. Relative expression levels of OsDREB1A and OsMYB4 genes were measured via RT-qPCR in 10-day-old seedlings of Thaibonnet and Volano grown under a long-day (14/10 h, day/night) photoperiod, and subsequently under control (28 °C/28 °C) and overnight cold (28 °C/10 °C) conditions. As shown in Fig. 1, RT-qPCR of OsDREB1A and OsMYB4 genes showed an upregulation under the 10 °C treatment overnight for both genotypes. Expression profiles of these two genes revealed that 2 and 10 h of plant exposure to 10 °C in the dark (which means 16 and 24 h after the beginning of the experiment) could be assumed as critical time-points for the “early” and “late” response in transcriptional activity.
Relative expression levels of OsDREB1A and OsMYB4 genes in Thaibonnet and Volano seedlings exposed to cold (28 °C/10 °C) under long-day photoperiod (14/10 h, light/dark) for two consecutive days. Data are expressed as relative units, assuming as reference the value of Thaibonnet at the beginning of the first day. Gray background indicates dark time
RNA-Seq data analysis
Total RNA of treated and control leaves from Thaibonnet (sensitive) and Volano (tolerant) plants was paired end-sequenced with Illumina HiSeq. Overall, the number of paired reads ranged from 24 to 43 million across the 18 RNA samples (Table 2), and after a quality check of the libraries with FastQC (Andrews 2010), adapters and low-quality nucleotides were filtered out. Filtered reads (90 to 94%, Table 2) were aligned to Oryza sativa ssp. Japonica (IRGSP-1.0 assembly) and Oryza sativa ssp. Indica (ASM465v1 assembly) genomes: as expected, the percentage of reads mapping to Japonica genome was higher than to Indica one for both Thaibonnet and Volano, confirming that both cultivars are genetically closer to Japonica than to Indica subspecies.
Among the 37,830 Oryza sativa ssp. Japonica annotated coding genes, the inactive ones in all the samples were filtered out, and thus, 22,376 active genes were analyzed for Thaibonnet and 22,747 for Volano. Normalization factor was calculated according to each library size (Online Resource 3); expression values of all transcribed genes are reported as EdgeR-normalized read counts on Online Resource 4. Online Resource 5 reports multi-dimensional scaling (MDS) plots for Thaibonnet and Volano libraries generated with EdgeR: this visualization of the differences between the samples expression profiles shows a very high reproducibility between biological replicates and a remarkable difference between samples.
Differential expression analysis
Normalized read counts of transcribed genes were used as input data for differential expression analysis, using EdgeR likelihood test: results for all genes were reported in Online Resource 6 and graphically represented in Fig. 2, while a list of genes that were differentially expressed in at least one of the 4 DE analysis is reported on Online Resource 7. As shown in Fig. 3 and Table 3, the number of DEGs were higher after 10 h of chilling stress than after 2 h. Moreover, the number of DEGs in Volano was slightly higher than in Thaibonnet. Common and specific DEGs up- and downregulated in the four genotype/stress conditions were also represented in the Venn diagrams of Fig. 4.
As evident in Fig. 3 and Table 3, upregulated genes in 2-h-stressed samples almost doubled the downregulated ones for both genotypes, while no significant differences were observed between the numbers of up- and downregulated genes in 10-h-stressed samples. This was mainly due to the higher number of commonly upregulated genes that was almost three times the downregulated one (1483 vs. 593) at 2 h.
To better observe the entity of “early” and “late” gene regulation in the two varieties, DEGs were divided in 6 clusters on the basis of their expression trend, and the distribution of log2 (fold change) at 2 and 10 h of stress was reported in Online Resource 8. While no relevant differences between the cultivars were observed in clusters of genes regulated at 2 or 10 h only, a trend of stronger modulation can be noticed in Volano for genes up- or downregulated in both stress conditions. In fact, for those clusters of genes regulated in both time points, expression changes were basically stronger in Volano than in Thaibonnet.
To confirm the reliability of RNA-Seq data, we evaluated the consistence between differential expression analysis and RT-qPCR results for OsMYB4 and OsDREB1A genes. As shown in Table 4, expression trends for OsDREB1A and OsMYB4 genes obtained with the two methods were coherent, and validated the RNA-Seq experiment for further data interpretation.
DEG distribution along rice genome and relationship with known QTLs
With the aim of better understanding the genetic bases of response to chilling, the relationship between DEGs and their physical position on the genome was investigated. The number of common and cultivar-specific DEGs for each mega-base along rice chromosomes was calculated, and normalized by the number of annotated genes in order to minimize the bias due to irregular gene distribution along the chromosomes (Mizuno et al. 2011). A map of DEG distribution over the rice genome was produced using these normalized frequency values (Fig. 5). Some chromosome regions (e.g., Chr 1: 35–40Mb; Chr 3: 5–10Mb; Chr 4: 25–30Mb; Chr 9: 15Mb-end; Chr 10: 20Mb-end) show a high frequency of genes that are differentially expressed under chilling stress in both genotypes. Moreover, several remarkable regions in which the density of specific DEGs sensibly diverge between Volano and Thaibonnet were observed (e.g., Chr 1: 30–35Mb; Chr 2: 20–25Mb; Chr 5: 20–25Mb; Chr 7: 20–25Mb; Chr 9: 0–5Mb; Chr 10: 15–20Mb; Chr 12: 20-25 Mb). These loci harbor groups of physically linked genes whose expression varies only (or mainly) in one of the two cultivars, indicating a possible role in chilling tolerance. To correlate the DEG distribution map with known variations in this trait phenotype, genomic regions related to cold tolerance at different developmental stages (including seedling and spike) and to other traits that could influence it (osmotic adjustment, cell membrane stability) were collected from Gramene database (www.gramene.org; Tello-Ruiz et al. 2016). Moreover, QTLs for cold tolerance at seedling stage not reported in Gramene were also added to the map: qCTS12 (Andaya and Tai 2006), qCTS4 (Andaya and Tai 2007), qCtss11 (Koseki et al. 2010), qSCT1 and qSCT11 (Kim et al. 2014), qLOP2 and qPSR2-1 (Xiao et al. 2015), COLD1 (Ma et al. 2015). Physical positions on rice genome of these QTLs were inferred by searching sequences of associated markers (including flanking and peak) by BLASTN in Os-Nipponbare-Reference-IRGSP-1.0 pseudomolecules (Online Resource 9). When overlapping, QTLs related to the same trait were merged and reported as unique loci in Fig. 5, so that interesting co-location with density peaks of common and genotype-specific DEGs could be identified (e.g., the distal part of Chr 1 long arm; Chr 6: 5–10Mb; Chr 8: 18–28Mb; Chr 9 long arm; Chr 11 short arm).
Frequency distribution map of common and genotype-specific DEGs. Frequency is expressed as number of DEGs for each chromosome Mb, normalized by the number of annotated genes in the same Mb. Common DEG frequency is represented with black lines, while red and blue ones stand for Thaibonnet and Volano specific DEG frequencies, respectively. Chromosomal position refers to the Oryza sativa ssp. Japonica IRGSP-1.0 pseudomolecules. QTLs associated to cold tolerance and to either membrane stability or osmotic adjustment capacity are reported on the left of each chromosome as white and gray bars, respectively
Gene ontology enrichment analysis
Gene ontology (GO) enrichment analysis of the four DEG sets was conducted to reveal biological processes differentiating the two contrasting genotypes. GO terms were considered as enriched if their p value was lower than a 0.05 threshold: a total of 407 GO terms were found to be enriched in at least one of the four genotype/stress combinations (Online Resources 10 and 11).
In total, 193 enriched GO terms were shared by both genotypes. Among the 95 GO terms related to Biological Processes (BP), some were consistent with responses to low temperatures stress, including “response to stress” (GO:0006950), “MAPK cascade” (GO:0000165), “regulation of defense response” (GO:0031347) and “stress-activated protein kinase signaling cascade” (GO:0031098). Other GO terms not directly attributable to chilling response, but whose importance could be relevant due to the linkages between response pathways to different abiotic stresses (Nakashima et al. 2014), such as “response to water deprivation” (GO:0009414) and “response to wounding” (GO:0009611), were found to be enriched in the transcriptomes of both genotypes. As far as genotype specific enriched gene ontology terms belonging to Biological Process (BP) class are concerned, 55 BP GO terms were enriched in Volano but not in Thaibonnet (Fig. 6). Among them, GO terms related to hormones such as “auxin efflux” (GO:0010315), “jasmonic acid metabolic process” (GO:0009694) and “response to gibberellin” (GO:0009739) resulted to be enriched only in Volano and only after 10 h of chilling stress. On the other hand, 67 GO terms were enriched in Thaibonnet (sensitive) but not in Volano (resistant). Some of these GO-terms related to hormones such as “cytokinin-activated signaling pathway” (GO:0009736), “cellular response to abscisic acid stimulus” (GO:0071215), “gibberellin catabolic process” (GO:0045487) and “abscisic acid binding” (GO:0010427) were enriched already at 2 h, while GO terms “response to water” (GO:0009415) and “response to salt stress” (GO:0009651) were enriched only after 10 h of chilling. These differences in enriched GO terms between contrasting genotypes indicate that pathways related to these terms could be differentially implicated in response to chilling stress in sensitive and resistant genotypes.
Discussion
Differential expression analyses in Thaibonnet and Volano after 2 and 10 h at low temperatures identified interesting short-term “early” and “late” transcriptional responses, and were compared to previous reports on rice response to chilling stress. The high number of DEGs found here (about 8500 in each genotype after 10 h of chilling), is in accordance with do Amaral et al. (2016), who studied the effects of 3 abiotic stresses (low temperatures, salinity and iron toxicity) on rice transcriptome and, interestingly, chilling was reported to modulate the highest number of genes. According to Zhang et al. (2014), rice is known to react to low temperature through three main signal cascades: an ICE-DREB1/CBF pathway, a MAPK cascade and an ABA-dependent pathway. Some aspects of the observed Volano and Thaibonnet response are discussed below in relation to these three cascades and other chilling-related pathways. For ease of reference, DE analysis results for the genes mentioned in this paragraph were extracted from Online Resource 7 and reported in Table 5.
ICE-CBF/DREB1 pathway
This response pathway has been widely studied in the model plant Arabidopsis since the end of 1990s and many of its components have also been identified in rice (Chinnusamy et al. 2007; Zhu et al. 2007; Zhang et al. 2013; do Amaral et al. 2016). The transduction cascade is triggered by a transient increase of cytosolic Ca2+, whose signal is integrated by various family proteins such as calcium-dependent protein kinases (CDPKs) (Asano et al. 2012) and calmodulin binding transcription activators (CAMTA) (Doherty et al. 2009). These kinases induce the activity of the basic helix-loop-helix transcription factor ICE (Inducer of CBF Expression), which in turn activates the key drought responsive element binding (DREB)/C-repeat/dehydration-responsive element binding factor (CBF) genes. DREBs/CBFs encode AP2/EREBP transcription factors that act as major “regulatory hubs” for downstream cold-regulated (COR) effector genes (Zhou et al. 2011).
In our analysis, transcription of many CDPK genes, such as OsCDPK4 (Os02g0126400), OsCDPK5 (Os02g0685900), and OsCDPK7 (Os04G0584600), was observed to be activated in both cultivars during chilling exposure. Expression of OsCDPK7 was already reported to be induced under salt and cold treatments (Saijo et al. 2000). Two other OsCDPKs seem to have a different behavior between sensitive and tolerant genotypes: OsCDPK21 and OsCDPK23 corresponding to Os08g0540400 and Os10g0539600, respectively. OsCDPK21 showed a striking increase (ca. 16-fold) of gene activity in Volano already after 2 h of stress, while a weaker and later response was observed in Thaibonnet (ca. 4-fold at 10 h). A similar trend (but with lower induction) was observed for OsCDPK23 (Table 5). Interestingly, these two genes had never been specifically reported as cold-responsive: OsCDPK21 was known to enhance tolerance to salt stress (Asano et al. 2011), while OsCDPK23 was previously reported as a protein kinase required for storage product accumulation during seed development (Asano et al. 2002). This differential transcript accumulation between contrasting rice cultivars suggest that these two calcium-dependent protein kinases might putatively be involved in tolerance to low temperatures. As firstly reported in Arabidopsis by Chinnusamy et al. (2003), CDPKs proteins activate the inducer of CBF expression (ICE1) transcription factor, a bHLH protein that acts as a positive regulator of CBF3 expression. The rice homolog of ICE1 gene (OsICE1), whose activity was demonstrated to improve photosynthetic performance under abiotic stresses (Chander et al. 2018), was studied by Nakamura et al. (2011), who observed that cold stress increased the levels of OsICE1 proteins, whereas no difference in gene expression was revealed. On the contrary, in our experiment, OsICE1 (Os01g0705700) was firmly upregulated equally at 2 and 10 h, although in Volano absolute values of transcription enhancement were slightly higher than in Thaibonnet. As far as OsDREB1 transcription factors are regarded, OsDREB1A (Os09g0522200), OsDREB1B (Os09g0522000), and OsDREB1H (Os09g0522100) activities peak at 2 h and have a log2FC of about 7 in Thaibonnet and 10 in Volano in our experiment, which means that in tolerant genotypes they are 8 times more induced than in sensitive ones. These results are coherent with the findings of Dubouzet et al. (2003) who observed that OsDREB1A and OsDREB1B are early induced upon chilling stress. However, while no alterations in OsDREB1C (Os06g0127100) activity were observed in their analysis, in our experiment, this latter gene was early-induced by low temperatures only in tolerant genotype with a log2FC of about 3. Noteworthy, in our experiment, OsDREB1G (Os02g0677300) had a peculiar behavior that could suggest its role in tolerance to chilling, besides drought (Chen et al. 2008). This gene was strongly upregulated after 2 h in both genotypes (log2FC of 3.30 in Thaibonnet and 5.86 in Volano) but then, while its expression fully decreases in Thaibonnet, it remained firmly active in Volano, possibly extending the COR genes activation period and, consequently, contributing to Volano’s major tolerance to chilling stress. MYB transcription factors are part of ICE-CBF/DREB1 signal cascade too, as some of them regulate DREB1/CBFs activity. In particular, OsMYB3R-2 (Os01g0841500) and OsMYB2 (Os03g0315400) were previously reported to increase low temperatures tolerance (Zhang et al. 2014) and, consistently, in our experiment we observed that those genes increased their activity after chilling stress. While OsMYB2 was sensibly activated at 2 h and decreased at 10 h, OsMYB3R-2 transcript was higher at 10 h stage only. OsMYBS3 (Os10g0561400) gene was upregulated since 2 h in the sensitive genotype, while just at 10 h in the tolerant one and, as it was reported to negatively regulate cold tolerance (Zhang et al. 2012, 2014), our results confirm its role in chilling response.
MAPK cascade
This pathway is triggered by reactive oxygen species (ROS) accumulation in cold-treated rice cells and, as reported by Xie et al. (2009), by a subsequent activation of a downstream cascade involving mitogen-activated protein kinase (MAPK), MAPK kinase (MAPKK) and MAPKK kinase (MAPKKK) genes. Plant MAPK are involved in signaling pathways induced by abiotic stresses, including low temperatures (Suarez-Rodriguez et al. 2010; Liu and He 2017). This signal cascade activates transcriptional regulation genes that promote the production of COR (cold-responsive) proteins (Sinha et al. 2011; Zhao et al. 2017). In particular, OsMKK6 (Os01g0510100), OsMPK3 (Os03g0285800), and OsMPK6 (Os10g0533600) were previously observed to increase cold stress tolerance when over-expressed in transgenic rice plants (Xie et al. 2012). In our experiment, OsMKK6 and OsMPK6 genes showed a weak “late” response (only in Volano for OsMPK6; Table 5), while OsMPK3 was induced immediately after exposure to low temperatures, according to the findings that this gene responds early to stress (Wang et al. 2013). On the other hand, OsTRX23, a cold-induced thioredoxin that negatively regulates kinase activity (Xie et al. 2009), was not differentially expressed in our experiment. Therefore, while confirming that OsMKK6, OsMPK6, and OsMPK3 genes are low-temperature responsive and thus supporting their involvement in the common part of the response mechanism, they do not seem to be determinant for the chilling tolerance difference between Volano and Thaibonnet. This is also supported by the enrichment of “MAPK cascade” GO term (GO:0000165) observed in both cultivars.
ABA-dependent pathway
This cold response pathway is triggered by abscisic acid (ABA) transient accumulation. Abscisic acid interacts with ABA-responsive genes via cis-acting ABA-response elements (ABRE) and ABRE-binding bZIP transcription factor (ABF) (Hossain et al. 2010). Then, OsNAC gene transduces the ABA signal through an AREB in its promoter and regulates the expression of NACRS-containing genes to increase cold tolerance in rice (Nakashima et al. 2007, 2012; Song et al. 2011). Many OsABF and bZIP genes were investigated in our experiment. However, they result poorly (Os06g0211200, Os02g0766700, Os01g0867300) or not differentially expressed at all (Os09g0456200, Os08g0472000, Os05g0489700, Os07g0686100). OsNAC4 (Os01g0816100) and OsNAC6 (Os01g0884300) genes have a stronger upregulation since early stage of low temperatures response, while OsNAC5 (Os11g0184900) was upregulated only at the late stage, and its induction was higher in Thaibonnet than in Volano. DREB1D is the only known DREB1/CBF gene involved in Arabidopsis ABA-dependent cold-response (Haake et al. 2002) and drought-tolerance (Guttikonda et al. 2014) pathways, but its rice orthologous (OsDREB1D, Os06g0165600) was not differentially expressed in our experiment. All this suggest that ABA-dependent pathway are not determinant for the differential response of the two cultivars. Such result was also confirmed by the enrichment of “ABA-activated signaling pathway” GO term (GO:0009738) common to Volano and Thaibonnet.
Summarizing DEG analysis for the three major cold-responsive pathways, a prominent role of the ICE-DREB1/CBF pathway in chilling tolerance mechanisms was revealed in Japonica rice like in other Poaceae (Akhtar et al. 2012; Miura and Furumoto 2013): the stronger upregulation of several OsDREB1 genes could explain phenotypic differences in tolerance among cultivars. Interestingly, transcription trends of some OsDREB1 genes (OsDREB1C, OsDREB1H and OsDREB1G) are novelties that expand our knowledge on the set of genes involved in chilling tolerance mechanism. Moreover, differential transcript accumulation of calcium-dependent protein kinases (OsCDPK21 and OsCDPK23) suggests their role in tolerance to low temperatures. Further research is needed to clarify their role at the molecular level and to unveil their specific contribution to chilling tolerance.
Hormonal cross-talk
In abiotic stress conditions, signal cascades modulate changes in transcriptional regulation, biosynthesis and transport of phytohormones (Munné-Bosch and Müller 2013). As mentioned above, a cold response pathway is known to be triggered by ABA accumulation (Zhou et al. 2011; Zhang et al. 2014), and changes in ABA generate non-linear regulations in the production of other phytohormones such as salicylic and jasmonic acids (da Maia et al. 2016). This “hormonal cross-talk” is confirmed by our results where, among the GO-terms enriched in both rice cultivars, several are related to biosynthesis, signaling and response to hormones such as ABA (GO:0009738), salicylic acid (GO:0009751, GO:0010337, GO:0009863, GO:0009697, GO:0080142), jasmonic acid (GO:2000022, GO:0009753, GO:0009867), auxin (GO:0009733, GO:0009851) and ethylene (GO:0009723). In particular, jasmonic acid (JA) is known to have a prominent role in abiotic stress response: its level in stressed rice is reported to be significantly higher than in non-stressed control (Du et al. 2013). Moreover, accumulation of JA was observed to be higher in abiotic stresses-tolerant rice genotypes than in sensitive ones (Kang et al. 2005). Our gene expression analysis confirmed these findings, since genes involved in JA biosynthesis responded to chilling stress: CYP74A2 (Os03g0225900) whose expression was enhanced in both genotypes, while CYP74A1 (Os03g0767000) was more upregulated in Volano. OsJAZ8 (Os09g0439200) was interestingly upregulated just in Thaibonnet and, since JAZ proteins are known to act as repressors of JA-responsive genes (Chung et al. 2008), OsJAZ8 could putatively repress the plant response to JA only in chilling-sensitive genotype. A role of JA in chilling tolerance was also highlighted by GO enrichment analysis: “Jasmonic acid biosynthetic process” GO-term (GO:0009695) was enriched in both genotypes for genes modulated after 2 h of chilling stress while, at 10 h, this GO category resulted enriched only in Volano (Online Resource 11).
Other contributors?
In addition to the genes involved in the processes described above, some other transcripts were found to have an interesting expression profile (Table 5), and their differential short-term response to cold in the contrasting genotypes Volano and Thaibonnet could suggest their involvement in conferring chilling tolerance to rice seedlings. Plant cytochrome P450 monooxygenases catalyze several enzymatic reactions for various metabolites (Heitz et al. 2012; Hamberger and Bak 2013) and some P450 genes were found to be upregulated in rice cold-tolerant genotypes (da Maia et al. 2016). In our experiment, genes belonging to P450 family did not show a diversified trend in expression between the contrasting genotypes but, interestingly, CYP76M8 (Os02g0569400) resulted strongly upregulated only in tolerant genotype (at both early and late stage). In the present study, also a heat shock protein similar to HSP70B (Os01g0688900) resulted intensely upregulated since the early stage of chilling stress exclusively in tolerant genotype as well: its expression trend is coherent with the already known involvement of heat shock proteins in response to cold (Zhang et al. 2012; Dametto et al. 2015). Noteworthy, a peroxidase gene (POX-1, Os04g0688200) was induced only in Volano at 2 h of stress, and its upregulation rose up to log2FC = 5.43 at 10 h, while in Thaibonnet no differential expression was observed. This result confirms the observations of Cheng et al. (2007) and could be explained with the role of peroxidases as ROS scavengers: resistant genotypes have an higher activity of this gene, whose product contribute to lower the concentration of ROS in stressed cells, conferring a higher level of tolerance to low temperatures. Expression profiles of mentioned genes, although interesting from the present research point of view, would need further molecular characterization to prove their role in chilling tolerance.
Early response matters
Cereals monitor temperature with a high level of precision, and differences in the initial rates of acclimation have been proposed as a fundamental mechanism differentiating tolerant and susceptible varieties (Fowler 2008). A demonstration of the faster rate of acclimation in resistant genotypes was provided by Fowler and Limin (2004), who linked differences in threshold temperatures and phenotypes by comparing a set of reciprocal near isogenic lines in wheat. From a molecular point of view, a similar approach was followed in the present study to compare “early” and “late” response in rice. Genes that were upregulated in both tolerant and sensitive genotypes, but whose activity grew up earlier in Volano than in Thaibonnet could be crucial: their early activation could in fact trigger faster acclimation in Volano and determine increased tolerance. Among the genes that were upregulated in both tolerant and sensitive genotypes, 249 resulted activated from 2 h after stress in Volano, but only after 10 h in Thaibonnet. MapMan software (Usadel et al. 2009) analysis showed that transcription factors were the most represented class among these genes (Online Resource 12). In particular, six helix-loop-helix DNA-binding domain containing proteins (bHLH, the same class as ICE1 gene) and three ethylene-responsive binding proteins (AP2/EREBP, same class of DREB transcription factors) (Online Resource 13) showed this faster upregulation in Volano. Such evidence confirms the importance of transcription factors for low-temperature response, and suggest their contribution in chilling tolerance.
DEG distribution and QTLs position
Analysis carried out on positional relationships between DEGs and previously identified QTLs revealed target regions that could be prioritized in breeding superior rice genotypes (Fig. 5). Some chromosomal segments were particularly dense of DEGs, showing their high frequency in both genotypes. A probable explanation is that these regions harbor groups of genes responsible for an important part of the chilling response mechanism, shared by tolerant and sensitive cultivars. Noteworthy, the density peak in the distal part of chromosome 9 long arm coincided with the physical position of DREB1/CBF genes cluster, where genes OsDREB1A, OsDREB1B, and OsDREB1H were found upregulated in both accessions, with higher induction in tolerant genotype. This cluster is conserved in Poaceae (Tondelli et al. 2011), and its structure suggests a tendency to gene duplication in the course of evolution: a greater number of genes belonging to DREB1/CBF family usually means an increased level of low-temperature tolerance (Francia et al. 2015, 2016). Some chromosomal regions in which the density of specific DEGs sensibly diverge between contrasting genotypes could also be observed (e.g., Chr 1: 30–35Mb; Chr 2: 20–25Mb; Chr 5: 20–25Mb; Chr 7: 20–25Mb; Chr 9: 0–5Mb; Chr 10: 15–20Mb; Chr 12: 20–25Mb). These loci putatively include groups of physically linked genes that respond to chilling only in one cultivar, so they can contribute to rice chilling tolerance mechanism. Notably, Volano specific DEGs density (blue lines in Fig. 5) overcome the Thaibonnet one (red lines) in most of these regions, which also suggests that DEGs in these loci are more often specific for the tolerant genotype. This observation is in line with DE analysis statistics in which the number of DEGs specific to Volano was higher than the number of Thaibonnet, especially for the early response (Fig. 3; Table 3). Our results also confirm the asymmetric distribution of active genes in the centromeric regions of the rice chromosomes (Mizuno et al. 2011) and, although more abundant in chromosome distal regions, active genes could be found close to centromeres as well. Some of these genes are induced by chilling stress and could have a role in tolerance mechanism. This could represent a hurdle during the breeding process, since the recombination frequency in pericentromeric regions is lower compared to telomeres and subtelomeres. In this case, recently developed techniques like CRISPR/Cas (Cong et al. 2013) could be a functional instruments to edit genes included in these regions, and to introduce positive or improved alleles in elite germplasm. For some genomic loci, high DEG frequency was correlated with QTLs known to be linked to cold tolerance and to other traits that could influence it (osmotic adjustment and cell membrane stability; e.g., the distal part of Chr 1 long arm; Chr 6: 10–20Mb; Chr 8: 18–28Mb; Chr 9 long arm, and Chr 11 short arm): these genomic regions putatively play an important role in low temperature response in rice. Some QTLs also corresponded to regions where density of Thaibonnet and Volano specific DEGs sensibly diverge (e.g., Chr 1: 30–35Mb; Chr 2: 20–25Mb; Chr 3: 30–35Mb; Chr 4: 25–25Mb; Chr 7: 20–25Mb). Besides above mentioned DREB1/CBF genes that correlates with a DEGs density peak, we observed other correspondence between known QTLs, DEGs density and noteworthy differentially expressed genes. For example, OsCDPK7 (Os04G0584600), OsCDPK5 (Os02g0685900), OsCDPK21 (Os08g0540400) and OsMYB3R-2 (Os01g0841500) were included respectively in AQDU015, qPSR2–1, COLD2 and CQP1 QTLs related to cold tolerance, and coincided with density peaks of common DEGs. Although no coincidence was found between OsMYB4 and the position of already known QTLs, OsMYB2 (Os03g0315400) coincided with a density peak of common DEGs that co-located with a QTL putatively responsible for osmotic adjustment capacity (AQFT003). These correspondences indicated rice genome loci that are putatively highly significant for chilling tolerance mechanism, and could possibly be used for future breeding programs.
Conclusions
In this study, we used an RNA-Seq approach to profile the short-term transcriptional response of Japonica rice to chilling. Most known low-temperature response pathways were analyzed and, interestingly, ICE-DREB1/CBF cascade resulted to play a primary role in differential response to chilling of the two contrasting cultivars Volano (tolerant) and Thaibonnet (sensitive). A clear involvement in tolerance mechanism of both known (OsICE1, OsDREB1A, OsDREB1B) and novel (OsDREB1C, OsDREB1G, and OsDREB1H) transcription factors was demonstrated. Transcriptional regulators were in fact the most represented class among the genes that were upregulated earlier in the tolerant than in the sensitive genotype: this evidence confirms the primary role of TFs in acclimation, and suggests their fast activation as a physiological mechanism leading to higher tolerance. Besides genes already known to be involved in chilling response, we found some novel genes that show a different expression trend between the two contrasting genotypes (i.e., calcium-dependent protein kinases OsCDPK21 and OsCDPK23, Cytochrome P450 monooxygenase CYP76M8, Peroxidase POX-1, etc.), and are putatively implicated in short-term low temperature response in rice. Differentially expressed genes density was calculated along all the rice chromosomes, and related to the QTLs associated to low temperature response. This co-localization of DEGs and QTLs on the chromosomes delivers a general overview of rice chromosomic regions that determine response to chilling in seedlings, and contributes to a better understanding of the molecular mechanisms underlying it. Moreover, GO enrichment analysis evidenced that phytohormones-related GO terms play a prominent role in both common and genotype-specific response, confirming their importance for low temperature response signaling. Overall, a deep reconfiguration of the transcriptome in both Volano and Thaibonnet succeeding the chilling stress was observed, and a great number of molecular mechanisms and signal transduction pathways were involved in these alterations. However, we described some important evidences that could provide an explanation for the differences between the contrasting genotypes: a stronger upregulation of several OsDREB1 genes in the tolerant genotype, a slower activation of some transcription factors in susceptible one, and an overlapping between known chilling tolerance-related QTLs and genomic regions where the density of cultivar-specific DEGs sensibly diverge. Our results provide a solid background for future development of chilling tolerant Japonica genotypes.
References
Akhtar M, Jaiswal A, Taj G et al (2012) DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet 91:385–395. https://doi.org/10.1007/s12041-012-0201-3
Andaya V, Mackill D (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theor Appl Genet 106:1084–1090. https://doi.org/10.1007/s00122-002-1126-7
Andaya VC, Tai TH (2006) Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet 113:467–475. https://doi.org/10.1007/s00122-006-0311-5
Andaya VC, Tai TH (2007) Fine mapping of the qCTS4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Breed 20:349–358. https://doi.org/10.1007/s11032-007-9096-8
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. https://doi.org/10.1093/bioinformatics/btu638
Andrews S (2010) Babraham Bioinformatics—FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Asano T, Kunieda N, Omura Y, Ibe H, Kawasaki T, Takano M, Sato M, Furuhashi H, Mujin T, Takaiwa F, Wu CY, Tada Y, Satozawa T, Sakamoto M, Shimada H (2002) Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell 14:619–628. https://doi.org/10.1105/tpc.010454
Asano T, Hakata M, Nakamura H, Aoki N, Komatsu S, Ichikawa H, Hirochika H, Ohsugi R (2011) Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol 75:179–191. https://doi.org/10.1007/s11103-010-9717-1
Asano T, Hayashi N, Kikuchi S, Ohsugi R (2012) CDPK-mediated abiotic stress signaling. Plant Signal Behav 7:817–821. https://doi.org/10.4161/psb.20351
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Boudsocq M, Laurie C (2005) Osmotic signaling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiol 138:1185–1194. https://doi.org/10.1104/pp.105.061275
Caffagni A, Pecchioni N, Francia E, Pagani D, Milc J (2014) Candidate gene expression profiling in two contrasting tomato cultivars under chilling stress. Biol Plant 58:283–295. https://doi.org/10.1007/s10535-014-0403-z
Chander S, Almeida DM, Serra TS, et al (2018) OsICE1 transcription factor improves photosynthetic performance and reduces grain losses in rice plants subjected to drought. Environ Exp Bot in press:. doi: https://doi.org/10.1016/j.envexpbot.2018.02.004
Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198. https://doi.org/10.1007/s10529-008-9811-5
Cheng C, Yun K-Y, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun S, de los Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics 8:175. https://doi.org/10.1186/1471-2164-8-175
Chinnusamy V, Ohta M, Kanrar S, Lee B-H, Hong X, Agarwl M, Zhu J-K (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev 17:1043–1054. https://doi.org/10.1101/gad.1077503
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451. https://doi.org/10.1016/j.tplants.2007.07.002
Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146:952–964. https://doi.org/10.1104/pp.107.115691
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
da Maia LC, Cadore PRB, Benitez LC, Danielowski R, Braga EJB, Fagundes PRR, Magalhães AM, Costa de Oliveira A (2016) Transcriptome profiling of rice seedlings under cold stress. Funct Plant Biol 44:419–430. https://doi.org/10.1071/FP16239
Dametto A, Buffon G, Blasi ÉA (2015) Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal … 2324:. doi: https://doi.org/10.1080/15592324.2015.1057369
do Amaral MN, Arge LWP, Benitez LC, Danielowski R, Silveira SFS, Farias DR, de Oliveira AC, da Maia LC, Braga EJB (2016) Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct Integr Genomics 16:567–579. https://doi.org/10.1007/s10142-016-0507-y
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. 21:972–984. doi: https://doi.org/10.1105/tpc.108.063958
Du H, Liu H, Xiong L (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4:397. https://doi.org/10.3389/fpls.2013.00397
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high salt and cold responsive gene expression. Plant J 33:751–763
Ente Nazionale Risi (2017) Superfici investite a riso 2016 - dati espressi in ettari. http://www.enterisi.it/upload/enterisi/bilanci/St1bis-1617_15916_366.pdf
FAO (2015) FAO Statistical Pocketbook 2015
Fowler DB (2008) Cold acclimation threshold induction temperatures in cereals. 48:1147–1154. doi: https://doi.org/10.2135/cropsci2007.10.0581
Fowler DB, Limin AE (2004) Interactions among factors regulating phenological development and acclimation rate determine low-temperature tolerance in wheat. Ann Bot 94:717–724. https://doi.org/10.1093/aob/mch196
Francia E, Pecchioni N, Policriti A, Scalabrin S (2015) CNV and structural variation in plants: prospects of NGS approaches. In: Advances in the understanding of biological sciences using next generation sequencing (NGS) approaches. Springer International Publishing, Cham, pp 211–232
Francia E, Morcia C, Pasquariello M, Mazzamurro V, Milc JA, Rizza F, Terzi V, Pecchioni N (2016) Copy number variation at the HvCBF4 HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol Biol 92:161–175. https://doi.org/10.1007/s11103-016-0505-4
Guttikonda SK, Valliyodan B, Neelakandan AK, Tran LSP, Kumar R, Quach TN, Voothuluru P, Gutierrez-Gonzalez JJ, Aldrich DL, Pallardy SG, Sharp RE, Ho THD, Nguyen HT (2014) Overexpression of AtDREB1D transcription factor improves drought tolerance in soybean. Mol Biol Rep 41:7995–8008. https://doi.org/10.1007/s11033-014-3695-3
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648. https://doi.org/10.1104/pp.006478
Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368. https://doi.org/10.1098/rstb.2012.0426
Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, Werck-Reichhart D, Pinot F (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287:6296–6306. https://doi.org/10.1074/jbc.M111.316364
Hossain MA, Il CJ, Han M et al (2010) The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167:1512–1520. https://doi.org/10.1016/j.jplph.2010.05.008
IRRI (2002) Standard evaluation system for rice. IRRI, The Philippines
Ji SL, Jiang L, Wang YH et al (2008) QTL and epistasis for low temperature Germinability in Rice. Acta Agron Sin 34:551–556. https://doi.org/10.1016/S1875-2780(08)60021-8
Kang DJ, Seo YJ, Lee JD, Ishii R, Kim KU, Shin DH, Park SK, Jang SW, Lee IJ (2005) Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J Agron Crop Sci 191:273–282. https://doi.org/10.1111/j.1439-037X.2005.00153.x
Kanneganti V, Gupta AK (2008) Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol 66:445–462. https://doi.org/10.1007/s11103-007-9284-2
Kim S-J, Lee S-C, Hong SK, An K, An G, Kim SR (2009) Ectopic expression of a cold-responsive OsAsr1 cDNA gives enhanced cold tolerance in transgenic rice plants. Mol Cell 27:449–458. https://doi.org/10.1007/s10059-009-0055-6
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. https://doi.org/10.1186/gb-2013-14-4-r36
Kim S-M, Suh J-P, Lee C-K, Lee JH, Kim YG, Jena KK (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Gen Genomics 289:333–343. https://doi.org/10.1007/s00438-014-0813-9
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Koseki M, Kitazawa N, Yonebayashi S, Maehara Y, Wang ZX, Minobe Y (2010) Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol Gen Genomics 284:45–54. https://doi.org/10.1007/s00438-010-0548-1
Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. https://doi.org/10.1371/journal.pcbi.1003118
Liu Y, He C (2017) A review of redox signaling and the control of MAP kinase pathway in plants. Redox Biol 11:192–204. https://doi.org/10.1016/j.redox.2016.12.009
Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J, Guo X, Xu S, Niu Y, Jin J, Zhang H, Xu X, Li L, Wang W, Qian Q, Ge S, Chong K (2015) COLD1 confers chilling tolerance in rice. Cell 160:1209–1221. https://doi.org/10.1016/j.cell.2015.01.046
Mackill DJ, Lei X (1997) Genetic variation for traits related to temperate adaptation of Rice cultivars. Crop Sci 37:1340. https://doi.org/10.2135/cropsci1997.0011183X003700040051x
Mao D, Chen C (2012) Colinearity and similar expression pattern of Rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One 7:e47275. https://doi.org/10.1371/journal.pone.0047275
Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, Zhang D, Chen C (2015) Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet 128:1359–1371. https://doi.org/10.1007/s00122-015-2511-3
Maruyama K, Urano K, Yoshiwara K, et al (2014) Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. 164:1759–1771. doi: https://doi.org/10.1104/pp.113.231720
Miura K, Furumoto T (2013) Cold signaling and cold response in plants. Int J Mol Sci 14:5312–5337. https://doi.org/10.3390/ijms14035312
Mizuno H, Kawahara Y, Wu J, Katayose Y, Kanamori H, Ikawa H, Itoh T, Sasaki T, Matsumoto T (2011) Asymmetric distribution of gene expression in the Centromeric region of Rice chromosome 5. Front Plant Sci 2:1–12. https://doi.org/10.3389/fpls.2011.00016
Munné-Bosch S, Müller M (2013) Hormonal cross-talk in plant development and stress responses. Front Plant Sci 4:1–2. https://doi.org/10.3389/fpls.2013.00529
Nakamura J, Yuasa T, Huong TT, Harano K, Tanaka S, Iwata T, Phan T, Iwaya M (2011) Rice homologs of inducer of CBF expression (OsiCE) are involved in cold acclimation. Plant Biotechnol 28:303–309. https://doi.org/10.5511/plantbiotechnology.11.0421a
Nakashima K, Tran L-SP, Van Nguyen D et al (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630. https://doi.org/10.1111/j.1365-313X.2007.03168.x
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103. https://doi.org/10.1016/j.bbagrm.2011.10.005
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170. https://doi.org/10.3389/fpls.2014.00170
Niroula RK, Pucciariello C, Ho VT, Novi G, Fukao T, Perata P (2012) SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J 72:282–293. https://doi.org/10.1111/j.1365-313X.2012.05078.x
Pan XW, Li YC, Li XX et al (2013) Differential regulatory mechanisms of CBF regulon between Nipponbare (japonica) and 93-11 (indica) during cold acclimation. Rice Sci 20:165–172. https://doi.org/10.1016/S1672-6308(13)60121-3
Payne R (2014) Regression, nonlinear and generalized linear models 88
Pecchioni N, Kosová K, Vítámvás P, Prášil IT, Milc JA, Francia E, Gulyás Z, Kocsy G, Galiba G (2014) Genomics of low-temperature tolerance for an increased sustainability of wheat and barley production. In: Genomics of plant genetic resources. Springer Netherlands, Dordrecht, pp 149–183
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327. https://doi.org/10.1046/j.1365-313X.2000.00787.x
Sato Y, Masuta Y, Saito K, Murayama S, Ozawa K (2011) Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep 30:399–406. https://doi.org/10.1007/s00299-010-0985-7
Shen C, Li D, He R, Fang Z, Xia Y, Gao J, Shen H, Cao M (2014) Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J Plant Biol 57:337–348. https://doi.org/10.1007/s12374-014-0183-1
Sinha AK, Jaggi M, Raghuram B, Tuteja N (2011) Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav 6:196–203. https://doi.org/10.4161/psb.6.2.14701
Song S-Y, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345. https://doi.org/10.1007/s00425-011-1403-2
Suarez-Rodriguez MC, Petersen M, Mundy J et al (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649. https://doi.org/10.1146/annurev-arplant-042809-112252
Tello-Ruiz MK, Stein J, Wei S, Preece J, Olson A, Naithani S, Amarasinghe V, Dharmawardhana P, Jiao Y, Mulvaney J, Kumari S, Chougule K, Elser J, Wang B, Thomason J, Bolser DM, Kerhornou A, Walts B, Fonseca NA, Huerta L, Keays M, Tang YA, Parkinson H, Fabregat A, McKay S, Weiser J, D'Eustachio P, Stein L, Petryszak R, Kersey PJ, Jaiswal P, Ware D (2016) Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res 44:D1133–D1140. https://doi.org/10.1093/nar/gkv1179
Tondelli A, Francia E, Barabaschi D, Pasquariello M, Pecchioni N (2011) Inside the CBF locus in Poaceae. Plant Sci 180:39–45. https://doi.org/10.1016/j.plantsci.2010.08.012
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Usadel B, Poree F, Nagel A et al (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, Maize. Plant Cell Environ 32:1211–1229. https://doi.org/10.1111/j.1365-3040.2009.01978.x
Wang Q, Li J, Hu L, Zhang T, Zhang G, Lou Y (2013) OsMPK3 positively regulates the JA signaling pathway and plant resistance to a chewing herbivore in rice. Plant Cell Rep 32:1075–1084. https://doi.org/10.1007/s00299-013-1389-2
Wang D, Liu J, Li C et al (2016) Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 9:61. https://doi.org/10.1186/s12284-016-0133-2
Wang Y, Jiang Q, Liu J, Zeng W, Zeng Y, Li R, Luo J (2017) Comparative transcriptome profiling of chilling tolerant rice chromosome segment substitution line in response to early chilling stress. Genes Genomics 39:127–141. https://doi.org/10.1007/s13258-016-0471-x
Xiao N, Huang W, Li A, Gao Y, Li YH, Pan CH, Ji H, Zhang XX, Dai Y, Dai ZY, Chen JM (2015) Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor Appl Genet 128:173–185. https://doi.org/10.1007/s00122-014-2420-x
Xie G, Kato H, Sasaki K, Imai R (2009) A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Lett 583:2734–2738. https://doi.org/10.1016/j.febslet.2009.07.057
Xie G, Kato H, Imai R (2012) Biochemical identification of the OsMKK6–OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J 443:95–102. https://doi.org/10.1042/BJ20111792
Yang Q-S, Gao J, He W-D, Dou TX, Ding LJ, Wu JH, Li CY, Peng XX, Zhang S, Yi GJ (2015) Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genomics 16:446. https://doi.org/10.1186/s12864-015-1551-z
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92. https://doi.org/10.1126/science.1068037
Yun K-Y, Park MR, Mohanty B, Herath V, Xu F, Mauleon R, Wijaya E, Bajic VB, Bruskiewich R, de los Reyes BG (2010) Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol 10:16. https://doi.org/10.1186/1471-2229-10-16
Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274. https://doi.org/10.1371/journal.pone.0043274
Zhang Q, Jiang N, Wang G-L, Hong Y, Wang Z (2013) Advances in understanding cold sensing and the cold-responsive network in Rice. Adv Crop Sci Tech 1:1–8. https://doi.org/10.4172/2329-8863.1000104
Zhang Q, Chen Q, Wang S, Hong Y, Wang Z (2014) Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice (N Y) 7:24. https://doi.org/10.1186/s12284-014-0024-3
Zhao X-Q, Wang W-S, Zhang F, Zhang T, Zhao W, Fu BY, Li ZK (2013) Temporal profiling of primary metabolites under chilling stress and its association with seedling chilling tolerance of rice (Oryza sativa L.). Rice (N Y) 6:23. https://doi.org/10.1186/1939-8433-6-23
Zhao C, Wang P, Si T, Hsu CC, Wang L, Zayed O, Yu Z, Zhu Y, Dong J, Tao WA, Zhu JK (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43:618–629.e5. https://doi.org/10.1016/j.devcel.2017.09.024
Zhou MQ, Shen C, Wu LH, et al (2011) CBF-dependent signaling pathway: A key responder to low temperature stress in plants. 31:186–192. doi: https://doi.org/10.3109/07388551.2010.505910
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290–295. https://doi.org/10.1016/j.pbi.2007.04.010
Zhu Y, Chen K, Mi X, Chen T, Ali J, Ye G, Xu J, Li Z (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in Rice. PLoS One 10:e0145704. https://doi.org/10.1371/journal.pone.0145704
Acknowledgements
Thanks are due to Marco Moretto and Paolo Sonego (Fondazione Edmund Mach, San Michele all’Adige ITALY) for their precious help with RNA-Seq data treatment.
Funding
This work was supported by Progetto AGER, grant n° 2010-2369—Integrated Genetic And Genomic Approaches For New Italian Rice Breeding Strategies (RISINNOVA).
Author information
Authors and Affiliations
Contributions
MP, DR, VTH, and CP performed rice plants phenotiping, RNA extractions and RT-qPCR. MB and JAM performed the RNA-Seq analysis. MB wrote the manuscript. EF, NP, and PP conceived the experiment, participated in the interpretation and discussion of results, and contributed to the writing of the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Key message
Short-term transcriptional response to chilling stress of rice seedlings was profiled. Novel genes implicated in tolerance mechanism were found, and relationships between gene expression profiles and known QTLs were reported, contributing to a better understanding of mechanisms underlying it.
Electronic supplementary material
Online Resource 1
Primers used for RT-qPCR. (XLSX 9 kb)
Online Resource 2
Statistical analysis of electrolytic leakage (EL%) measured in not stressed plants (ctrl) and 2, 7, 14 days after chilling exposure for Experiment 1. Dashed lines inside boxes represent the average value for the 13 cultivars. Significant differences were derived from unbalanced ANOVA and reported on the figure (n.s., not significant; ***, p < 0.001). (PNG 32 kb)
Online Resource 3
Size (bp) and normalization factors (calculated with EdgeR according to each library size) for the 18 RNA-Seq libraries. (XLSX 11 kb)
Online Resource 4
Normalized reads counts for active genes in Thaibonnet and Volano RNA libraries. (XLSX 6378 kb)
Online Resource 5
Multi-dimensional scaling (MDS) plots for Thaibonnet (a) and Volano (b) RNA libraries generated with EdgeR to measure gene expression differences (three biological replicates for 0, 2 and 10 h of chilling stress for each genotype). (PNG 29 kb)
Online Resource 6
EdgeR differential expression analysis results for all the active genes of Thaibonnet and Volano rice genotypes for 2 and 10 h of chilling stress compared to control samples. (XLSX 10721 kb)
Online Resource 7
List of the rice genes that are differentially expressed in at least one of the four stress combinations (Thaibonnet/Volano; 2/10 h of stress) compared to controls. Position on the genome, description and log2 Fold Change resulting from EdgeR analysis are reported for each gene. Empty cells on log2FC columns mean that the gene is not regulated on the correspondent RNA-Seq set. (XLSX 1011 kb)
Online Resource 8
Box plots representing the log2(Fold Change) statistical distribution for DEGs of Thaibonnet (red) and Volano (blue), clustered by their expression trend: (a) genes up-regulated after 2 and 10 h of stress; (b) genes up-regulated only after 2 h of stress; (c) genes up-regulated only after 10 h of stress; (d) genes down-regulated after 2 and 10 h of stress; (e) genes down-regulated only after 2 h of stress; (f) genes down-regulated only after 10 h of stress. Whiskers above and below the boxes indicate the 10th and the 90th percentiles. (PNG 296 kb)
Online Resource 9
List of QTLs found in Gramene and in bibliography. For each QTL, relative trait, linkage group (LG), genetic position in cM (where available), associated markers, physical position in Japonica rice genome in bp, number of genes included in the QTL that are differentially expressed in at least one experiment (Thaibonnet/Volano and 2/10 h) are reported. (XLSX 17 kb)
Online Resource 10
GO-enrichment analysis results for DEGs of Thaibonnet and Volano rice genotypes for 2 and 10 h of chilling stress. For each GO term, over- and under-represented p-value, number of DE genes (numDEInCat), total genes (numInCat), ontology and term name are included. (XLSX 1190 kb)
Online Resource 11
List of the GO terms enriched (p < 0.05) in at least one of the 4 analyzed DEGs sets (Thaibonnet/Volano and 2/10 h). For each GO term, p-value for each DEGs set, ontology and GO term name are reported. Empty cells on p-value columns means that the GO term is not significantly enriched in the correspondent DEGs set. (XLSX 40 kb)
Online Resource 12
MapMan regulation overview of the genes that are up-regulated at 2 h in Volano, but only at 10 h in Thaibonnet. Genes are binned to MapMan functional categories and the values are represented as the log2-transformed values. Blue squares indicates up-regulated genes. (PNG 79 kb)
Online Resource 13
Genes binned by MapMan to “Transcription Factor” among genes that are up-regulated at 2 h and 10 h in Volano, but only at 10 h in Thaibonnet. (XLS 37 kb)
Rights and permissions
About this article
Cite this article
Buti, M., Pasquariello, M., Ronga, D. et al. Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp. Japonica seedlings. Funct Integr Genomics 18, 627–644 (2018). https://doi.org/10.1007/s10142-018-0615-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0615-y