Abstract
14-3-3s are a class of conserved proteins in eukaryotes involved in almost all cellular processes. Published studies have shown that this family plays a role in response to stress conditions. In this study, comparative genomics identified thirty-one 14-3-3 cDNAs encoding 25 unique proteins with transcripts that were sufficiently divergent in the coding regions to define different 14-3-3 members. Phylogenetic analysis indicated that the cotton 14-3-3 family was similar to the family in the plant model species Arabidopsis and rice. The multiple copies of 14-3-3 genes in cotton are suggested to be the result of a recent duplication event after the divergence of cotton from its progenitor species. 14-3-3s are differentially expressed in a range of cotton organs as well as under different environmental conditions. Salinity and drought stress significantly induced the altered expression of 14-3-3 genes, suggesting that 14-3-3s play a role in cotton response to environmental abiotic stress. The 14-3-3 gene CGF14-4 was more sensitive to both drought and salinity stress with a 10.2-fold change under 1% of PEG treatment, while other 14-3-3s responded only to either drought or salinity stress. These results implied that specific isoforms of 14-3-3s play different regulatory roles in stress response besides their role in development. 14-3-3s were more sensitive to abiotic stress in roots than in leaves, suggesting that the root is more sensitive to stress treatment than leaves. The differential expression pattern of 14-3-3s also suggests the occurrence of functional divergence among specific isoforms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton is one of the most important industrial crops because of its natural textile fiber and edible oil. The huge economic value of cotton drives scientists to improve the economic character of cotton, such as its fiber quality and yield at different environmental conditions, including stresses. Among the four cultivated species of Gossypium, upland cotton (Gossypium hirsutum) is the most widely planted species and accounts for about 95% of world cotton production (Chen et al. 2007). Therefore, upland cotton is most often the focus of research among all of the cotton species. G. hirsutum is allopolyploidy (AADD, 2n = 4x = 52), and the haploid genome size (x = 13) is estimated to be approximately 2.5 Gb (Chen et al. 2007; Hendrix and Stewart 2005). The genome size and the complexity of cotton have hindered whole genome sequencing (Chen et al. 2007), but studies from different research groups have accumulated over 268,786 expressed sequence tag (EST) sequences in GenBank.
In almost all eukaryotic species, 14-3-3 genes exist as multiple isoforms, with two in yeast, seven in most mammals, and more than a dozen in plants (van Heusden 2005). Monocotyledon rice (Oryza sativa) contains eight genes, and each of them is expressed (Chen et al. 2006; Yao et al. 2007). Dicotyledons such as Arabidopsis thaliana and soybean (Glycine max) have 15 and 18 genes, with 12 and 16 shown to be expressed (Rosenquist et al. 2001), respectively. The 14-3-3 protein sequences are about 250-amino acid long and usually form homodimers or heterodimers. Up to now, 14-3-3 proteins have been reported to interact with diverse proteins, over 300 proteins in Arabidopsis, by regulating the activity of the proteins in phosphoserine-dependent and phosphoserine-independent manners (Chang et al. 2009), and thus are involved in almost all known cellular process as regulators, such as signal pathways, transcriptional regulation, protein subcellular localization, development, and stress response (van Heusden 2005).
Although high sequence similarity exists among multiple copies of 14-3-3s, the protein isoform specificity has been implicated under the observation of different subcellular localization, tissue-specific expression, and dynamic regulation in response to environmental change (Kjarland et al. 2006). It has been demonstrated that several 14-3-3 genes are involved in response to environmental stress in tobacco (Nicotiana tabacum; Chen et al. 1994), A. thaliana (Jarillo et al. 1994), and rice (O. sativa; Chen et al. 2006). Currently, nine cDNAs encoding 14-3-3 proteins have been reported in cotton (Shi et al. 2007; Zhang et al. 2010), two of which may be involved in salt stress response (Wei et al. 2009). Interestingly, the constitutive expression of the Arabidopsis 14-3-3 lambda isoform in transgenic cotton enhances the tolerance to salt stress (Yan et al. 2004). However, there is no systematical study on identifying the transcripts of 14-3-3s in cotton; as a result, there have been no systematical investigations on the expression and function of 14-3-3 genes under stress conditions. Here, we systematically identified 14-3-3 genes in cotton at the genome-wide level by assembling the ESTs using a rigorous method, and we also investigated the gene expression of the representative members of this gene family under salt and drought stress.
Materials and methods
Identification of 14-3-3 genes
The homolog identification of 14-3-3 genes was performed by two rounds of BLAST searching. In the first round of BLAST searching, pre-assembled ESTs of G. hirsutum using the Contig Assembly Program (CAP3; Huang and Madan 1999) were downloaded from the cotton EST database (http://www.leonxie.com) and were used as a local database; protein sequences annotated as “14-3-3” were downloaded from GenBank and were used as queries. All of the hits with score values more than 50 were retrieved and were used as queries in the second round of BLAST searching against the GenBank non-redundant (nr) database. Only the genes with 14-3-3 genes as the top hits were kept as candidates.
The reassembly of the ESTs for 14-3-3 genes was performed using MIRA (Chevreux et al. 2004) in two different ways based on the results of the pre-assembled ESTs by CAP3. These two ways differ in using two different datasets as the input files of MIRA. One way is to use all of the ESTs in each contig of GAP3, and in this case, MIRA was run for each of contig. Another way is to use all of the ESTs of all the contigs of GAP3. After obtaining these two assembly results of MIRA, we manually checked each of them using SeqMan gene analysis software (DNASTAR Inc., Madison, WI, USA). Only the consensus sequences supported by no less than two ESTs were kept for further detailed analyses. For the consensus sequences that still contain more than one gene, we carried out manual assignment for the polymorphism sites. Because we reassembled them two times using the same two datasets, the resulting consensus sequences contain multiple redundant sequences. To remove the redundancy of assembly, we considered the following two aspects: (1) out of consensus sequences in which 100% identities were present with each other, the longer one was kept and (2) consensus sequences derived from the same ESTs were checked carefully and only the reliable sequence was retained. To make sure that each of the assembled sequences comes from real genes, we also excluded the shorter sequences that show less than two nucleotide site differences in the coding region with the corresponding ones. The coding region (CDS) and untranslated region (UTR) were determined by blastx against the GenBank nr database and the CDSs were manually retrieved with ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). All of the consensus sequences and their corresponding ESTs are available on our web site (http://www.leonxie.com).
Phylogenetic analysis
To investigate protein isoform diversity of cotton 14-3-3s, phylogenetic analysis was conducted on the putative cotton protein sequences and the homologs from A. thaliana, O. sativa, and humans. Because 6 of the 31 cDNAs obtained by our cDNA assembly show 100% identity in amino acid sequences, only the 25 divergent protein sequences were used in our phylogenetic analysis. Other sequences were retrieved from GenBank using their accession numbers published in previous studies (Li and Dhaubhadel 2011). Multiple sequence alignments were performed using ClustalX (Thompson et al. 1997), and gaps and ambiguously aligned sites were removed manually. The JTT model with four gamma distribution rates was chosen as the most appropriate protein substitution matrix by ModelGenerator (Keane et al. 2006). A maximum likelihood method using PHYML (Guindon and Gascuel 2003) was used to conduct the phylogenetic analysis with 100 pseudo-replicates.
Cotton treatments and sample collection
The seeds of G. hirsutum cv.TM-1 were collected and hulled before surface sterilization. The seeds were then treated with 70% (v/v) ethanol for 60 s, 6% (v/v) bleach for 6–8 min, and sterile water washed four times. The sterilized seeds were grown on 1/2 Murashige and Skoog medium (pH 5.8) containing 0.8% agar under a 16-h light/8-h dark cycle at 24°C for 5 days. For salt and drought treatments, the medium was supplemented with 0.1%, 0.25%, 0.5%, and 1% NaCl and 1%, 2.5%, and 5% PEG, respectively. Roots and leaves were collected from the sterile seedlings and immediately stored in liquid nitrogen and then stored at −80°C until RNA extraction.
RNA isolation and real-time RT-PCR analysis
Total RNA was isolated from the grinded samples using the mirVanaTM miRNA Isolation Kit (Ambion, Austin, TX, USA). The procedure followed the manufacturer’s protocol, except that a sonication step for 15–20 s was added just after lysis of the cells. The quantity and quality of total RNA were assessed using a Nanodrop ND-1000 (Nanodrop Technologies, Wilmington, DE, USA). The mirVanaTM miRNA Isolation Kit is designed for small RNA extraction as well total RNA extraction; it has been well used in our lab for extracting great quality of both mRNAs and small RNAs (Frazier et al. 2011; Zhang et al. 2008). Quantitative real-time RT-PCR (qRT-PCR) was used to identify the expression patterns of selected 14-3-3 gene members in different cotton tissues (leaves and roots) under different stress conditions (salinity and drought). Firstly, reverse transcription was carried out using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and a poly-T tail primer. Secondly, qRT-PCR and gene-specific primers (Table 2) were used to verify the expression of selected genes. All the reactions were run on an Applied Biosystems 7300 Sequence Detection System according to the manufacturer’s protocol. The expression of selected14-3-3 genes were normalized using the mean value of eight commonly used housekeeping genes: eIF-5, histone 3, actin, UBQ7, ubiquitin, cydophilin, GAPDH, and alpha-tubulin. The expression levels were calculated using the ∆∆C t method. Three biological replicates were run for each gene at each stress treatment condition. Analysis of variance of single factor was used to compare the expression differences of the selected 14-3-3s under different salt and drought treatments in leaves and roots.
Results
Identification and nomenclature of 14-3-3 gene family in G. hirsutum
To find all potential 14-3-3 genes in cotton, we used all the currently available 1,472 sequences of 14-3-3 proteins in GenBank (as of December 10, 2010) as queries to search against the assembled EST database by GAP3 (Fig. 1). Among the 28,432 contigs in G. hirsutum that include 25,371 for consensus and 3,061 for singletons, 30 contigs were found with score values more than 50, and all of them were used as queries to search the GenBank nr database to check whether they really encode 14-3-3 genes. This second round of BLAST search showed that 19 of them hit 14-3-3 genes with high similarities. We viewed several assembled contigs and found that almost all of them contain three or more polymorphic sites representing two or more different transcripts supported by at least two ESTs (Fig. 2), which indicates that each of them should come from different real mRNAs rather than due to sequencing errors.
Fragment of one assembled contig by GAP3 showing three polymorphic sites (263, 274, and 300). The three polymorphic sites in red present two patterns, one is “T,” “A,” and “A”; the other is “C,” “G,” and “G”. Each of them is supported by multiple EST sequences. This indicates that these transcripts represent real mRNAs as they are produced in G. hirsutum rather than sequencing errors
Considering that GAP3 might have less power in the assembly of sequences with polymorphic sites, we utilized MIRA, which can split contigs into transcripts based on single-nucleotide polymorphisms, to reassemble these ESTs using recommended settings. We tried the assembly in two different ways. One is based on contigs resulted from GAP3. In this way, we only split each contig of CAP3 into specific transcripts using the ESTs in each contig as input files of MIRA. The other way is to use all the ESTs for 14-3-3 genes as the input file of MIRA. As for each assembly result of these two ways, we manually reassembled every specific transcript with SeqMan software and removed the fragments that might be due to sequencing error or come from less than two ESTs, which produced 35 and 39 contigs, respectively. Because these two assembly methods use the same dataset, the following two steps were adopted to eliminate the repeat-specific transcripts: (1) for the consensus sequences with 100% identities, the longer ones were retained and (2) for the consensus sequences assembled from the same ESTs, we carefully rechecked the assembly and only kept the reliable ones. Thus, we obtained 37 consensus sequences that exhibit polymorphic sites in the whole cDNA region.
To ensure that the consensus sequences are from different gene entities, the following points were considered. (1) Because the non-coding regions of mRNAs from one gene might present variant sites, only the coding regions of these 37 consensus sequences were compared and five pairs of sequences were found possessing 100% identities or only one nucleotide difference. Therefore, these five sequences were removed due to the possible post-transcription processing and sequencing error in ESTs. (2) Two sequences are partial and correspond to the 5′ and 3′ ends, respectively, and multiple sequence alignments show that they are closely related and might be from one transcript. Blast searches against the original ESTs fortunately revealed one EST sharing both of the polymorphic sites. Therefore, these two partial sequences were merged into one. (3) Finally, we obtained 31 consensus sequences with each of them representing one real mRNA (Table 1). Given that 24 of 31 cDNAs are full-length cDNAs, these mRNA probably are not derived by alternative splicing. Six pairs of these 31 cDNAs show 100% identity in the amino acid level. This means that these 31 cDNAs code for 25 different proteins.
Currently, the 14-3-3 genes in one species are named in alphabetical order if they represent real gene loci in the haploid genome. Nine mRNA sequences of G. hirsutum, obtained from cDNA libraries, have been submitted to GenBank and are named 14-3-3a to 14-3-3h and 14-3-3L. We compared our 31 mRNAs with the nine previously reported sequences. The result indicated that 9 of the 31 sequences show 100% identities in the nucleotide level with the corresponding ones. Given that the nine public mRNAs show relative high divergence and no indels are found within them, they are unlikely to be a result of alternative splicing or different allele genes but rather come from different gene loci. Therefore, it is reasonable to name them by following existing nomenclature for constancy. Each of five genes—14-3-3d, 14-3-3e, 14-3-3f, 14-3-3h, and 14-3-3L—has one additional cDNA sequence that is divergent in the nucleotide level but not in the amino acid level. Numbers were added to the current names to reflect the 100% identity with each other, such as 14-3-3d1 and 14-3-3d2. The remaining 17 mRNAs were named CGF14_1-16. Among them, CGF14_3.1 and CGF14_3.2 were named to indicate their shared 100% identity in the amino acid sequences.
Phylogenetic analysis of 14-3-3 gene family in G. hirsutum
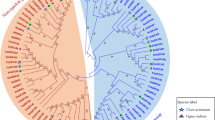
To investigate the sequence divergence and the potential functional difference of cotton 14-3-3 proteins, we performed phylogenetic analysis with the reported homologs from A. thaliana and O. sativa, which may represent dicots and monocots. Previous phylogenetic analysis on 14-3-3 proteins from Dictyostelium, plants, and animals demonstrated that this family has ε and non-ε two distinct groups in plants (Ferl et al. 2002). These two groups are also revealed in our phylogenetic tree (Fig. 3). The non-ε group includes at least two subgroups for each of the three species. The non-ε group presents three subclades, and subclade II contains more than one branch in cotton and only one branch in A. thaliana and O. sativa. To investigate the functional divergence of the new discovered 14-3-3 genes, we chose six genes in each of the subgroups as representatives to evaluate the response of cotton 14-3-3 genes to salt and drought stress in young cotton seedlings by qRT-PCR. Two previously reported genes encoding 14-3-3L and 14-3-3f were also investigated.
Molecular phylogenetic tree of 14-3-3 genes. The sequences in cotton are indicated in red letters. Sequence names beginning with AtGF and OsGF indicate that they are from A. thaliana and O. sativa, respectively. The red dots after the cotton sequence names indicate the gene expression profile by qRT-PCR performed in this study
Transcriptional profile of selected cotton 14-3-3 genes under salt and drought stress
Because of the high sequence similarity among the 31 cotton 14-3-3 genes, it is hard to design gene-specific primers in each cluster, although efforts had been made. We designed four newly identified genes in this study and also used two previously reported primer pairs encoding 14-3-3f1 and 14-3-3L1 (Table 2). Inspection of the potential amplification products showed that four of the six primer pairs—14-3-3L1, CGF14_4, CGF14_5, and CGF14_10—are gene-specific, and the other two primer pairs designed to amplify 14-3-3f1 and CGF14_13 may amplify 14-3-3f2 and CGF13_12, respectively. The two products amplified by 14-3-3f1/f2 only show 1°C difference in the melting temperature (T m), which exceeds the detection limit of this method and only presents one peak in the dissociation curves (data not shown). As expected, the products amplified by CGF14_12/13 show two peaks, and each product amplified by the other four primer pairs show one peak in the melting curves (data not shown).
The expression of 14-3-3 genes in cotton
Except 14-3-3f1, all of the other detected 14-3-3 genes showed higher expression levels than those of the reference gene. Among the highly expressed 14-3-3 genes, 13-3-3L1 is 1.5- to 2-fold that of the reference gene, while CGF14_4, CGF14_8, CGF14_10, and CGF14_14 exhibit near 50-fold or more that of the reference gene. For the individual 14-3-3 genes, CGF14_13 expresses higher in leaves than that in roots, and the expression of 14-3-3f1 and 14-3-3L1 in roots is about 1.5-fold that in leaves; the expression of the other 14-3-3 genes in roots is more than 6-fold that in leaves (Fig. 4).
14-3-3 genes response differently in leaves and roots under salinity and drought stress
In leaves, the expression of 14-3-3f1/f2, 14-3-3L1, CGF14_4, and CGF14_10 under salt stress declines with the increase of NaCl concentration (Fig. 5); the expression of CGF14_5 and CGF14_12/13 increases under low NaCl concentration and reaches the control level in high NaCl concentration, with the highest transcription level at 0.25% NaCl in CGF14_5 and at 0.1% NaCl in CGF14_12/13. The expression of CGF14_10 is significantly downregulated in leaves under 0.5% NaCl concentration compared with those under low NaCl concentration. Under drought stress, the expression of CGF14_10 is significantly upregulated in leaves under 1% PEG concentration. The expression of other selected 14-3-3 genes also increases, except that the expression of 14-3-3f1/f2, CGF14_4, and CGF14_10 is inhibited at certain PEG concentrations, with 14-3-3f1/f2 at medium PEG concentrations (2.5%), CGF14_4 at high PEG concentrations (5%), and CGF14_10 under medium and high PEG concentrations while increased under other PEG concentrations. The expression of CGF14_12/13 in medium PEG concentrations (2.5%) is inhibited, although it increases under salt stress.
In roots, all of the 14-3-3 genes exhibit increased expression levels (Fig. 5). However, the expression of 14-3-3f1/f2 and 14-3-3L1 is inhibited under high NaCl (0.5%) and high PEG concentrations (5%), the expression of CGF14_12/13 is inhibited under medium NaCl concentration (0.25%), and the expression of CGF14_5 under high PEG concentrations and CGF14_13 under low PEG concentrations is also inhibited.
Roots are more sensitive than leaves under salt and drought stress
Salt and drought treatments result in more significantly enhanced expression of all the studied 14-3-3s in roots than in leaves (Fig. 5). Under high salinity conditions, the largest fold change in roots were observed in CGF14_10 with a 13.1-fold upregulation after exposure to 0.25% NaCl; under drought conditions, the largest fold change was observed in CGF14_4 with a 10.2-fold upregulation at 1% PEG concentrations. However, the highest fold change in leaves under salt stress was observed in CGF14_13 with a 2.6-fold upregulation at the 0.1% concentration. Under drought stress, the largest 3.8-fold change in leaves was observed in CGF14_10 after being exposed to 1% PEG. In total, CGF14_10 shows a 29-fold change in roots than that in leaves under the same salt stress conditions and shows about a 14-fold change in roots than that in leaves under the same drought stress conditions.
The expression of 14-3-3 genes is dose-dependent under salt and drought stress
The expression changes of 14-3-3 genes in roots and leaves were dose-dependent and exhibited one of two trends in roots as the increase of PEG concentrations in the media (Fig. 5): (1) an increase in expression at low or medium PEG concentrations followed by a decrease in expression at high PEG concentrations or (2) a decrease in expression at medium PEG concentrations between an increase in expression in low and high PEG concentrations. 14-3-3 genes that fell into the first category included 14-3-3f1/f2, 14-3-3L1, CGF14_5, and CGF14_10 under salinity and drought conditions, and CGF14_4 under drought conditions, with the largest fold change at low PEG concentrations in CGF14_4 and CGF14_4 and the largest fold change at medium NaCl or PEG concentrations in other genes.
Discussion
Multiple copies of 14-3-3 transcripts and their evolutionary history in cotton
Although no cotton genome sequencing data are available, the abundant resource of ESTs allows us to investigate the transcripts of 14-3-3 gene family at the transcriptome level. Using two EST assembly softwares, manual inspection, and a rigorous standard to determine real transcripts with polymorphic sites, we identified 31 transcripts with divergence in protein coding regions. Some of these 31 transcripts may represent different gene loci, some may come from different allelic genes, and some may stand for different alternative splicing transcripts of the same gene.
Generally, the gene number within one species is determined by calculating how many genes are present in the haploid genome. Because the cotton genome contains duplication of large DNA segments within homolog chromosomes (Wang et al. 2007) and the identical genes from different gene loci would only present one transcript, it is hard to quantify the total gene number of 14-3-3s in this species with complex genome organization. However, the current data have not hindered us from making an approximate estimate on the gene number of this family since the identical transcripts from different gene loci most likely come from very recent gene or DNA segment duplication and therefore exhibit similar or identical transcriptional patterns.
Considering that G. hirsutum is allopolyploidy (AADD), we speculate that the obtained 31 cDNAs might come from the A and D genomes, with each of them containing about 16 transcribed genes. This is almost exactly consistent with the 16 transcribed genes in the soybean, G. max. The six pairs of cDNAs that show 100% identity in the amino acid level may indicate that sequence divergence has not happened between the genomes A and D. Investigation of diploid cotton, such as Gossypium arboretum, Gossypium herbaceum, or Gossypium raimondii, would help us verify this speculation.
Although cotton contains multiple transcript copies of 14-3-3s, our phylogenetic analysis indicated that the evolutionary history of this family is very similar to those of the homologs in A. thaliana and O. sativa, including recent gene duplication within each subgroups after the split of these three species. The additional one branch in subclade II of the ε group may imply potential novel functional specialization in cotton.
Different response properties of 14-3-3s in the transcript level reflect the occurrence of function divergence within this family
14-3-3s regulate many cell processes including stress response and plant nutrient signaling (Ferl et al. 2002; Shin et al. 2011). Although the important regulatory roles of 14-3-3 have been recognized, the isoform specificity has not been fully realized given the high identity among 14-3-3s. It is reported that the fine tuning in plants exposed to stress conditions often involves expression of specific 14-3-3 isoforms (Shin et al. 2011). The transcript levels of 14-3-3b and 14-3-3c were shown to increase in roots under salinity conditions in a previous study (Wei et al. 2009). Whether the other identified 14-3-3 genes show similar expression patterns, especially in different tissues, is unknown.
In this study, we used NaCl and PEG to simulate salinity and drought conditions, respectively, and investigated the expression level of 14-3-3 genes in different tissues under stress environments. The occurrence of functional divergence among 14-3-3 isoforms can be reflected in the following three aspects. Firstly, the expression of specific isoforms under stress conditions varies with different tissues. For example, expression of 14-3-3f1/f2, 14-3-3L1, and CGF14_10 is inhibited in leaves at the three tested salinity conditions, while it is enhanced in roots in most salinity conditions. Secondly, the expression of different isoforms changes in the same tissue. For example, in roots, at least two trends were observed under drought conditions. Thirdly, the expression of 14-3-3 genes alters in a dosage-dependent manner especially in drought conditions. The expression changes of 14-3-3 genes suggest their roles in differential regulation under salinity and drought conditions. Studies on their subcellular localization and target partners may facilitate our understanding about the role of this family in cotton and other organisms.
References
Chang IF, Curran A, Woolsey R, Quilici D, Cushman JC, Mittler R, Harmon A, Harper JF (2009) Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9:2967–2985
Chen F, Li Q, Sun L, He Z (2006) The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13:53–63
Chen Z, Fu H, Liu D, Chang PF, Narasimhan M, Ferl R, Hasegawa PM, Bressan RA (1994) A NaCl-regulated plant gene encoding a brain protein homology that activates ADP ribosyltransferase and inhibits protein kinase C. Plant J 6:729–740
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape JM, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S, Wilkins T, Wright RJ, Van Deynze A, Zhu Y, Yu S, Abdurakhmonov I, Katageri I, Kumar PA, Mehboob Ur R, Zafar Y, Yu JZ, Kohel RJ, Wendel JF, Paterson AH (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145:1303–1310
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WEG, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Research 14:1147–1159
Ferl RJ, Manak MS, Reyes MF (2002) The 14-3-3s. Genome Biol 3:REVIEWS3010
Frazier TP, Sun GL, Burklew CE, Zhang BH (2011) Salt and drought stresses iInduce the aberrant expression of microRNA genes in tobacco. Molecular Biotechnology. doi:10.1007/s12033-12011-19387-12035
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hendrix B, Stewart JM (2005) Estimation of the nuclear DNA content of Gossypium species. Ann Bot 95:789–797
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Jarillo JA, Capel J, Leyva A, Martinez-Zapater JM, Salinas J (1994) Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol Biol 25:693–704
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29
Kjarland E, Keen TJ, Kleppe R (2006) Does isoform diversity explain functional differences in the 14-3-3 protein family? Curr Pharm Biotechnol 7:217–223
Li X, Dhaubhadel S (2011) Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233:569–582
Rosenquist M, Alsterfjord M, Larsson C, Sommarin M (2001) Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol 127:142–149
Shi H, Wang X, Li D, Tang W, Wang H, Xu W, Li X (2007) Molecular characterization of cotton 14-3-3L gene preferentially expressed during fiber elongation. J Genet Genomics 34:151–159
Shin R, Jez JM, Basra A, Zhang B, Schachtman DP (2011) 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett 585:143–147
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
van Heusden GPH (2005) 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life 57:623–629
Wang K, Guo W, Zhang T (2007) Detection and mapping of homologous and homoeologous segments in homoeologous groups of allotetraploid cotton by BAC-FISH. BMC Genomics 8:178
Wei XZ, Zhang ZT, Li Y, Wang XL, Shao SQ, Chen L, Li XB (2009) Expression analysis of two novel cotton 14-3-3 genes in root development and in response to salt stress. Prog Nat Sci 19:173–178
Yan J, He C, Wang J, Mao Z, Holaday SA, Allen RD, Zhang H (2004) Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol 45:1007–1014
Yao Y, Du Y, Jiang L, Liu JY (2007) Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J Biochem Mol Biol 40:349–357
Zhang BH, Pan XP, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhang ZT, Zhou Y, Li Y, Shao SQ, Li BY, Shi HY, Li XB (2010) Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot 61:3331–3344
Acknowledgements
This work was supported partially by the USDA and the Cotton Incorporated (10-753).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, G., Xie, F. & Zhang, B. Transcriptome-wide identification and stress properties of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). Funct Integr Genomics 11, 627–636 (2011). https://doi.org/10.1007/s10142-011-0242-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0242-3