Abstract
High levels of water-soluble carbohydrates (WSC) provide an important source of stored assimilate for grain filling in wheat. To better understand the interaction between carbohydrate metabolism and other metabolic processes associated with the WSC trait, a genome-wide expression analysis was performed using eight field-grown lines from the high and low phenotypic tails of a wheat population segregating for WSC and the Affymetrix wheat genome array. The 259 differentially expressed probe sets could be assigned to 26 functional category bins, as defined using MapMan software. There were major differences in the categories to which the differentially expressed probe sets were assigned; for example, probe sets upregulated in high relative to low WSC lines were assigned to category bins such as amino acid metabolism, protein degradation and transport and to be involved in starch synthesis-related processes (carbohydrate metabolism bin), whereas downregulated probe sets were assigned to cell wall-related bins, amino acid synthesis and stress and were involved in sucrose breakdown. Using the set of differentially expressed genes as input, chemical–protein network analyses demonstrated a linkage between starch and N metabolism via pyridoxal phosphate. Twelve C and N metabolism-related genes were selected for analysis of their expression response to varying N and water treatments in the field in the four high and four low WSC progeny lines; the two nitrogen/amino acid metabolism genes demonstrated a consistent negative association between their level of expression and level of WSC. Our results suggest that the assimilation of nitrogen into amino acids is an important factor that influences the levels of WSC in the stems of field-grown wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Remobilisation of non-structural carbohydrate stored in the wheat stem maintains the supply of carbon to the grain when the rate of photosynthate production is less than the needs of the grain (Schnyder 1993) and makes a significant contribution to final grain yield and grain weight (Gebbing et al. 1999; van Herwaarden et al. 1998a, 2003; Rebetzke et al. 2008; Rattey et al. 2009). This carbon reserve is commonly referred to as water-soluble carbohydrates (WSC) and is composed predominantly of fructans of the graminan type (Chalmers et al. 2005). The accumulation and remobilisation of fructans in plants involves the concerted action of several enzyme families, namely fructosyltransferases (FT) and fructan exohydrolases (FEH). FTs play a key role in fructan biosynthesis whereas FEHs are usually involved in the degradation of fructan (van den Ende et al. 2003). Both spring and winter wheat lines have been shown to vary significantly for WSC concentration (WSCc) and WSC content on an area basis in stems around anthesis (Foulkes et al. 2002; van Herwaarden et al. 2003; Ruuska et al. 2006; Yang et al. 2007). WSC levels in wheat stems have also been shown to vary with water and N levels (McGrath et al. 1997; van Herwaarden et al. 1998a, b, 2003).
We have previously compared wheat lines with high or low levels of stem WSC and shown that higher expression levels of genes encoding fructan synthetic enzymes in the stem were correlated with increased stem WSC and fructan concentrations (Xue et al. 2008b) while mRNA levels of genes involved in sucrose hydrolysis were inversely correlated with WSC concentration. High WSC lines, compared to low WSC lines, also had lower mRNA levels of enzymes involved in diverting UDP-glucose to cell wall synthesis with a corresponding reduction in cell wall polysaccharide content, mainly hemicellulose, in the stem (Xue et al. 2008b). Expression levels of FEH genes have also been shown to correspond to variation in fructan levels. Van den Ende et al. (2003) observed that 1-FEH activity increases as fructans are mobilised from the stems with a subsequent gain in grain weight. Furthermore, Yang et al. (2004) noted that FEH genes co-located with QTL for grain weight, WSC and grain filling rate. More recently, Zhang et al. (2009) cloned the three 1-FEH genes and identified one homoeologue which is postulated to be the major gene contributing to FEH activity to facilitate fructan degradation and contribute to grain filling.
Expression of carbohydrate metabolic genes has also been shown to vary with water stress and level of N. Under water stress, levels of expression in the leaf of most genes encoding chloroplast enzymes involved in carbon fixation (Calvin cycle) were reduced whereas genes encoding cytoplasmic and vacuolar enzymes in the pathways leading to glucose, fructose and fructan production were upregulated (Xue et al. 2008a). Similarly, low N conditions resulted in increased fructan levels in young barley leaves and in wheat stems at anthesis via elevated levels of a suite FTs and 6-FEH (Wang et al. 2000; Ruuska et al. 2008) and stimulated other sucrose-utilising pathways, including components of starch biosynthesis. Ruuska et al. (2008) also identified several genes as potential regulators of C storage in stems, based on the correlation between fructan accumulation and levels of transcripts of these genes, including a vacuolar processing protease and kinases postulated to have a role in regulating C storage in starch.
In this paper, we have used global expression and chemical–protein interaction analyses to investigate the molecular mechanisms that control levels of WSC in wheat and in particular, the interaction between C and N metabolism gene expression and WSC accumulation in wheat. Using field-grown material from the phenotypic tails of a wheat population segregating for WSC, we have examined which genes were differentially expressed between high and low WSC lines and were responsive to environmental factors (nitrogen level and soil water status) that affect WSC accumulation. Using the set of differentially expressed genes as input, we have used chemical–protein network analysis; our results suggest that the conversion of nitrogen into amino acids controls the levels of WSC in stems of field-grown wheat.
Materials and methods
Germplasm, environments and phenotypic measurements
The Seri/Babax (SB) population (Olivares-Villegas et al. 2007) was evaluated across eight environments in northeastern Australia over the period 2002 to 2006, as described in Rattey et al. (2009). Genotype means from the 2002 to 2003 trials at Gatton (27°34′ S, 152°20′ E) in southeastern Queensland were used to identify eight high and eight low lines for WSC concentration, designated the “high” and “low” groups, respectively. The mean WSCc for these two groups over six environments at Gatton from 2002 to 2006 are shown in Fig. 1. The SB progeny within each group were selected such that the WSC group means were relatively similar for both days to anthesis and for plant height so as not to confound differences in WSC levels with these traits (data not shown).
Agronomic data were obtained in 2005 and 2006 from three trials conducted at Gatton. In 2005, the trial was mainly rainfed, while in 2006 there were both rainfed and irrigated trials. These three trials also included two nitrogen treatments (−N or +N) resulting in six managed environments across the three trials. Each of the three trials was a spatially arranged randomised complete block design with two replicates where the genotype × nitrogen combinations were randomised within each replicate. Details of these three trials are presented in Table 1. In all six managed environments, standard crop management practices were followed and crop measurements were taken as described by Rattey et al. (2009). Protein percentage in grain samples from plot harvests was measured by near-infrared reflectance spectroscopy (Crop Scan, NIR Technology Australia). Protein levels in biomass samples were estimated by multiplying the N concentration by 6.25 (van Herwaarden et al. 1998a; Dreccer et al. 2009). Structural biomass was estimated as the biomass less the amount of protein and amount of WSC; it was converted to a percentage by dividing by the amount of total biomass.
Statistical analysis of the phenotypic data across the six managed environments was undertaken using a two-stage process as outlined by Welham et al. (2006) and applied by Rattey and Shorter (2010) using customised script within the R software system (R Development Core Team 2006). Across environment BLUEs were estimated because the total G × E was small and the WSC_class × environment interaction was not significant (data not shown).
Generation, characterisation and analysis of gene expression data
Array and MapMan analysis
Fourteen of 16 hybridised Affymetrix GeneChip Wheat Genome Arrays prepared previously (Xue et al. 2008b—GEO Series number GSE9767) were re-analysed using GeneSpring GX 7.3.1 (Agilent Technologies, Santa Clara, CA, USA). The raw signal intensity values of the 61,127 probe sets present on the Affymetrix GeneChip Wheat Genome Array were pre-processed on import using RMA. The intensity value for each probe set was divided by the median of its measurements in all samples. The resulting normalised intensity values were filtered to remove probe sets for which no value exceeded 100. The entire dataset was also subjected to statistical analysis using one-way ANOVA with respect to WSC (α = 0.05, multiple testing correction: Benjamini and Hochberg FDR), to identify probe sets that were statistically significantly different in the two WSC groups. The intersection of these two data sets resulted in the final list of probe sets, whose expression was significantly different for WSC status and had intensity values of at least 100 for at least one sample.
For each probe set, the 14 relative intensity values from each array were examined to determine whether the probe set was upregulated or downregulated in the high WSC lines relative to the low WSC lines. The probe set was deemed to be upregulated in the high WSC group (U) if at least six from seven intensity values corresponding to the high WSC lines and no more than one value corresponding to the low WSC lines were greater than one and vice versa for probe sets deemed to be downregulated in the high WSC group (D).
The final list of probe sets with their WSC regulation status was imported into MapMan 3.0.0 (Thimm et al. 2004; Usadel et al. 2005) together with the mapping file containing the annotation of the Affymetrix GeneChip Wheat Genome Array (Alex Nagel, personal communication) to allow for the display of the data set across a variety of metabolic processes.
Interaction analysis
The Representative Public ID was retrieved for each probe set using NetAffx database at Affymetrix, Inc (http://www.affymetrix.com/). Using these as input, unique corresponding wheat TC and singleton DNA sequences were sourced from the DFCI Wheat Gene Index Release 12.0 (released on 18 April 2010—http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=wheat). Potential homologous sequences from Arabidopsis thaliana were identified using BLASTx of the TAIR10 Proteins at The Arabidopsis Information Resource (http://www.arabidopsis.org). The locus names, bit scores and E values of the best match to each wheat sequence were collated, and the resulting data set was filtered by requiring the E value to be 0.00001 or less in order to remove trivial matches. Duplicate locus names were removed prior to network generation. An interaction map was generated using STITCH 2.0 (http://stitch.embl.de/—Kuhn et al. 2010), with the A. thaliana locus list as input. A text file describing the network was downloaded and then visualised using Cytoscape 2.8.1 (http://www.cytoscape.org—Smoot et al. 2011).
Expression analysis in high and low WSC lines grown under high and low N and with and without irrigation
Stem samples were collected at anthesis from eight of the 16 SB lines grown in the six managed environments at Gatton in 2005 and 2006, as described in Xue et al. (2008b); these eight genotypes were the same as those used in the Affymetrix GeneChip Wheat Genome Array experiments. RNA was extracted and cDNA synthesised as previously described (Xue et al. 2008b). Real-time PCR was performed using two wheat genes, TaRPII36 (RNA polymerase II 36 kDa subunit) and TaRP15 (RNA polymerase I, II and III, 15 kDa subunit) as internal reference genes and 12 selected wheat genes (primer sequences for the wheat genes and for the two internal reference genes are listed in Supplementary Table 1) as described in Xue et al. (2008b). Statistical analysis of the expression data within and across environments was undertaken as described earlier in relation to the phenotypic data.
Results
WSC group, nitrogen and water effects on phenotype
Four high and four low WSC lines grown at Gatton in 2005 and 2006 in rainfed and irrigated trials that also featured nitrogen treatments were sampled for gene expression studies. To relate gene expression to agronomic performance of the same plant material, analyses were conducted on 13 agronomic traits for the four high and four low WSC lines grown in these six managed environments. Although grain yield in the six environments varied from 202 to 660 g/m2 and WSCc varied from 82 to 223 mg/g, G × E was small and the WSC_genotype group × environment interaction was not significant (data not shown). Differences between the high and low WSC groups in this restricted data set (Tables 2 and 3) were highly significant (P < 0.01) for WSCc and WSCt, culm number and weight, grain weight, grain number and percent grain protein (Tables 2 and 3), significant (0.01 < P < 0.05) for height and N% in anthesis biomass and non-significant for the remaining four traits (Table 2 and 3). When compared to the low WSC group, the high WSC group was slightly taller, had more WSC (concentration and content per culm), heavier but fewer culms, fewer but heavier grains and a higher percent grain protein (Tables 2 and 3). In addition, the high WSC genotype group had a lower structural biomass content, 71% versus 74% (data not shown), than the low WSC genotype group.
The interaction of WSC group with nitrogen was generally non-significant in the six managed environments so that differences between the WSC groups were unaffected by nitrogen level for most traits (data not shown). Across the six managed environments, the addition of nitrogen fertiliser increased the nitrogen percentage in anthesis biomass and percent grain protein but lowered WSCc (by 12%) (Tables 2 and 3); there were no significant differences for the other agronomic traits.
In the 2006 trials, there was no interaction of WSC group with water treatment (data not shown). The irrigated managed environment in 2006 had higher anthesis biomass, N% in anthesis biomass and taller plants but lower WSCc and WSCt than the rainfed trial (Table 2). At maturity, the irrigated managed environment had higher biomass, grain yield, grain number, culms per unit area and individual culm weight than the rainfed managed environment (Table 3). Grain weight was not significantly different between the two water regimes (Table 3) although there was a trend for heavier grain in the rainfed trial.
Genome-wide scan to identify differentially expressed genes in high versus low WSC lines
A total of 259 probe sets were identified whose expression was significantly different between the high and low WSC progeny lines. Of the 259 differentially expressed probe sets, 147 were deemed to be upregulated probe sets (U list—expressed more highly in the high WSC lines than in the low WSC lines), and 112 were downregulated probe sets (D list—less abundant in the high WSC lines than in the low WSC lines) (Supplementary Table 2).
The 259 annotated probe sets together with their differential expression status were imported in MapMan to provide a preliminary indication of the metabolic processes which are differentially regulated in high and low WSC lines. The 259 probe sets were mapped across 263 bin entries (four probe sets were each mapped to two bin entries) and were found to be distributed across 26 of the 36 available bins (Supplementary Table 2). The number of probe sets assigned to a bin with an ascribed function (i.e. not including bin 35—no assigned function) was compared between the U and D lists (Fig. 2). Differences in numbers, expressed as a percentage of the total, were evident between the two lists for most categories, and some bins were exclusively represented in either the U or D list. Categories that were represented exclusively in the U list were minor carbohydrate metabolism (bin 3), oxidative phosphorylation (bin 7), TCA/organic transformation (bin 8), N metabolism (bin 12), metal handling (bin 15), hormone metabolism (bin 17) co-factor and vitamin metabolism (bin18), redox (bin 21), nucleotide metabolism (bin 23), DNA (bin 28), development (bin 33) and transport (bin 34). In addition, RNA (bin 27, transcriptional regulation) and protein (bin 29, protein synthesis, degradation and modification) were more than three-fold higher in the U list when compared to the D list. The only category that was represented exclusively in the D list was cell wall (bin 10); however, the categories of amino acid metabolism (bin 13), secondary metabolism (bin 16), stress (bin 20), C1 metabolism (bin 25), miscellaneous enzymes (bin 26) and signalling (bin 30) were more than three-fold enriched in the D list when compared to the U list. Major carbohydrate metabolism (bin 2) displayed similar percentages for both sets of individuals. However, the probe sets in this bin that were upregulated in the high WSC lines relative to the low WSC lines corresponded to genes involved in starch metabolism while those downregulated in the high WSC lines relative to the low WSC lines corresponded to genes involved in the breakdown of sucrose (Supplementary Table 2).
MapMan bin membership for probe sets that were differentially expressed between high and low WSC wheat progeny lines. The probe sets in each bin and their regulation status are described in Supplementary Table 1. The bin numbers and their corresponding bin name are graphed on the y-axis. Percentage of probe sets in each bin is graphed on the x-axis
Interaction between C and N metabolism
The 259 differentially expressed probe sets were used as input for potential network identification using STITCH 2.0, a chemical–protein interaction database (Kuhn et al. 2010). The only information pertaining to plants in this database relates to A. thaliana, necessitating the identification of A. thaliana homologues to the identified wheat sequences. After sourcing the 238 unique corresponding wheat TCs and singletons, identification of the best match A. thaliana homologue and E value filtration to remove trivial matches, a set of 197 probe sets corresponding to 157 unique Arabidopsis loci was returned (Supplementary Table 2). This list of genes served as input to the STITCH 2.0 database, and an interaction map was generated using six active prediction methods—Neighbourhood, Gene Fusion, Co-occurrence, Co-expression, Experiments and Databases—to determine members of the potentially interacting gene set that interacted with each other and various metabolites at the highest confidence level (≥0.900). This map was then visualised using Cytoscape 2.8.1 (Smoot et al. 2011) in order to clarify the interactions, with interactions containing two or more input sequences being examined further (Fig. 3). The entire interaction map can be seen in Supplementary Figure 1 and the list of interactors in Supplementary Table 3.
Interaction maps of A. thaliana homologues of wheat transcripts differentially regulated between high and low WSC lines. White circles: input loci; pink circles: predicted interacting proteins (Supplementary Table 3); green squares: predicted interacting metabolites. Blue line: protein–protein interaction; green line: protein–metabolite interaction; red line: metabolite–metabolite interaction. Additional interaction networks with only one input locus are presented in Supplementary Fig. 1
Thirty-two input sequences were involved in interaction networks at the highest confidence level. Nineteen of the 32 input sequences were involved in three interaction networks containing two or more of the input sequences. One major interaction network containing genes with acknowledged roles in amino acid, nitrogen, C1, nucleotide and redox metabolism was identified. Four additional sub-networks containing genes involved in starch synthesis, protein synthesis, flowering induction and lipid metabolism, respectively, were attached to this major network via major metabolites. The lipid metabolism sub-network was also attached via a protein–protein interaction. The interaction between the starch synthesis sub-network and the main network was mediated by pyridoxal phosphate, while sulphate mediated interaction with flowering induction and adenosine diphosphate mediated interaction with both lipid metabolism and protein synthesis. Two additional small interaction networks were also visualised, one containing genes with known involvement in secondary metabolism and the other with an as-yet unknown role.
Effect of varying nitrogen and water on expression of genes in high and low WSC lines
As combined analyses showed that there were no significant two- or three-way interactions between WSC groups and water and/or N (data not shown), main effect differences of WSC group, N level or water level on gene expression levels for 12 genes in high and low WSC lines were assessed. As indicated in Table 4, the 12 selected genes encode (a) major enzymes involved in fructan synthesis (Ta1-SST1 and Ta6-SFT1), (b) potential regulators of the fructan synthetic pathway (TaVPE_, TaCBLipk1, TaWPK4), (c) enzymes involved in sucrose hydrolysis (TaSAInv1, TaSuS3, TaSuS5), (d) enzymes involved in diverting carbon into cell wall synthetic pathways (TaUGDH1, TaUGDC1) and (e) enzymes involved in nitrogen assimilation and amino acid synthesis (TaNIR1, TaGd). Most of these genes were differentially expressed between the high and low WSC lines (Supplementary Table 2, Xue et al. 2008b), or had been shown to be responsive to N levels and expressed in tissue actively synthesising WSC (Ruuska et al. 2008) (TaNIR1, TaVPE_, TaWPK4).
The results of the expression analyses are summarised in Table 5. For each of the 12 genes, the effect of WSC group (high or low WSC progeny), of level of Nitrogen applied (low or high levels of N) and of water (rainfed or irrigated trial in 2006) is indicated and significant differences noted. All 12 genes were strongly differentially expressed between high and low WSC lines across the six managed environments (Table 5). Ta1-SST1 and Ta6-SFT1 were more highly expressed in high WSC lines that in low WSC lines in contrast to both sucrose synthase genes and TaSAInv1 which were less expressed in the high WSC lines. The three regulators, TaVPE_, TaCBLipk1 and TaWPK4, were more strongly expressed in high WSC relative to low WSC lines. However, TaUGDH1 and TaUGDC1 were less expressed in the high WSC lines as were the nitrogen/protein-related genes TaNIR1 and TaGd.
Seven of the 12 genes were differentially expressed in response to N level across all lines (Table 5). Four of the five major carbohydrate metabolism-classified genes (see Tables 4 and 5) were not differentially expressed in response to N; Ta1-SST1 was differentially expressed with increased expression under low N conditions. Both cell wall metabolism-classified genes also were differentially expressed in response to N with increased expression under low N conditions. Four of the five nitrogen/protein metabolism/photosynthesis-classified genes were differentially expressed in response to N levels in the present study; the putative regulator TaWPK4 was not differentially expressed in response to level of N. Expression levels of TaNIR1, TaCBLipk1 and TaGd was greater in the high N conditions and lower for TaVPE_γ (Table 5).
Only five of the 12 genes were differentially expressed in response to water across all lines in the irrigated and rainfed trials of 2006 (Table 5): two carbohydrate metabolism-classified genes (Ta6-SFT1, TaSuS3), one cell wall metabolism-classified gene (TaUGDH1) and two nitrogen metabolism/photosynthesis-classified genes (TaNIR11, TaGd). None of the putative regulators was differentially expressed in response to water. Of the five genes that were differentially expressed in response to water, only expression of TaSuS3 was greater under rainfed (dry) conditions; expression of the other four genes was greatest in irrigated conditions. Only expression levels of TaNIR11 and TaGd showed a consistent inverse relationship with WSC across genotypes and all environmental conditions (Table 5).
Discussion
Multi-environment phenotypic assessment of high and low WSC lines
The phenotypic assessment of the high and low WSC groups across the six managed environments was consistent with results obtained in the full Seri–Babax population (Rattey et al. 2009) and in other populations and germplasm (van Herwaarden et al. 2003; Duggan et al. 2005; Ruuska et al. 2006; Rebetzke et al. 2008; Dreccer et al. 2009) in response to moisture levels (Ruuska et al. 2006) and N levels (van Herwaarden et al. 2003; Ruuska et al. 2006, 2008; Rebetzke et al. 2008). High WSC lines were associated with heavier grain weight and fewer culms per unit area, but not necessarily higher yield.
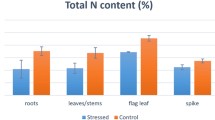
Both WSCc and WSCt were lower in this study than reported in other wheat populations (Rebetzke et al. 2008) but biomass N% was much higher. Of particular interest was the finding that WSC level around anthesis had a consistent negative association with biomass nitrogen percentage in varying N and water status environments that affect stem WSC accumulation. Across all lines, both low nitrogen and rainfed environments resulted in higher levels of WSCc and WSCt in wheat stems at anthesis but lower levels of biomass nitrogen than high nitrogen and irrigated environments, respectively; the high WSC SB lines also had a lower level of biomass nitrogen. These data are consistent with earlier studies (van Herwaarden et al. 1998a, b, 2003; Ruuska et al. 2008) and clearly imply an inverse relationship between WSC and nitrogen anabolism in wheat stems around anthesis; these observations prompted further study into genes underlying the WSC trait.
Different gene categories are differentially expressed between high and low WSC lines
The 259 probe sets representing genes that were differentially expressed between the high and low WSC lines could be assigned to 26 functional category bins using MapMan 3.0.0 (Thimm et al. 2004; Usadel et al. 2005) to categorise the probe sets. Some bins contained genes that were exclusively up- or downregulated in high versus low WSC lines, while other bins contained similar percentages of genes for both sets of lines but different sub-sets of genes were affected.
The functional category of several bins containing genes that were differentially expressed between high and low WSC lines is consistent with results reported in previous publications. Other studies have also noted that the expression of genes encoding enzymes with a major involvement in starch synthesis and degradation, such as those assigned to the major carbohydrate metabolism bin, were upregulated in high WSC lines relative to low WSC lines while enzymes involved in the breakdown of sucrose had lower levels of expression in high relative to low WSC lines (van den Ende et al. 2003; Yang et al. 2004; Ruuska et al. 2008; Xue et al. 2008b); these results indicate that there is considerable flux in starch accumulation in the stem. Although fructan is the predominant form of stored carbon in wheat stems, small amounts of starch also accumulate in wheat stems and leaves (Judel and Mengel 1982; Schnyder 1993; Scofield et al. 2009). Starch accumulation appears to occur earlier than WSC accumulation, from before heading to after anthesis, and is quickly re-utilised (Scofield et al. 2009). The timing, lesser amount and rapid re-utilisation of starch in the wheat stem suggest that the starch provides a temporary carbon supply for use in either active growth processes or for grain filling prior to the accumulation of fructan (Scofield et al. 2009).
Similarly, Xue et al. (2008b) had previously noted that high WSC wheat lines had lower levels of the two cell wall metabolism enzymes evaluated, UGDH1 and UGDC1. In the present study, numerous additional genes encoding enzymes with acknowledged roles in cell wall metabolism also had significantly lower levels of expression in high relative to low WSC lines. These genes included genes encoding enzymes involved in cell wall integrity and strength and synthesis (such as cell wall invertase, sucrose synthase and starch degradation enzymes which would assist to increase the hexose pool that would be available for cell wall synthesis). These observations are also consistent with differences in phenotype measured in the high and low WSC lines in the present study. Although biomass at both anthesis and maturity were similar in the two sets of lines, the low WSC lines had more structural biomass per unit area and a higher proportion of structural biomass per culm; similar results were found with a subset of these lines in other field experiments in which water availability, sowing date and/or N level were manipulated (Dreccer et al. 2009) and in other wheat populations (Rebetzke et al. 2008).
Few studies have examined the expression of genes involved in other metabolic categories in high and low WSC lines, including the expression of genes involved in N metabolism. In this study, the expression of genes encoding enzymes involved in amino acid and protein synthesis, such as those involved in phenylalanine (arogenate dehydratase—Yamada et al. 2008), methionine (methionine synthase MS—González-Verdejo et al. 2008; S-adenosylmethionine synthetase—Bhuiyan et al. 2007) and serine/glycine synthesis (serine hydroxymethyl transferase-SHMT and glycine decarboxylase-Gd—Bauwe and Kolukisaoglu 2003) were downregulated in the high WSC lines relative to the low WSC lines while the expression of genes encoding enzymes involved in protein degradation, such as numerous proteases, protein kinases and peptidases, was upregulated in high WSC lines; Ruuska et al. (2008) also reported increased expression levels of several kinases under low N conditions. These observations are consistent with the significantly lower concentrations of biomass nitrogen in the high WSC lines than in the low WSC lines in this and other studies (van Herwaarden et al. 1998b, 2003; Ruuska et al. 2008). In addition, known stress responsive genes and signalling genes were also downregulated in the high WSC lines. Interestingly, the expression of numerous transcription factors and transporters were upregulated in the high versus low WSC lines. In general, more probe sets were upregulated in the high WSC lines relative to the low WSC lines.
Interaction between carbohydrate metabolism and amino acid metabolism
There is considerable interaction between C and N metabolism in plants to ensure efficient assimilation of these two essential nutrients results in optimum plant development, growth and yield (Nunes-Nesi et al. 2010). Regulation of N and C assimilation and metabolism results from regulation of the transcription of genes involved in these processes as well as sensory systems to monitor C and N metabolism metabolite levels and post-transcriptional control by microRNAs (Gutierrez et al. 2007; Nunes-Nesi et al. 2010). The reduction of nitrate and ammonium, the main sources of N for many plants, requires energy from photosynthesis, and C skeletons from sucrose are required for the incorporation of N into molecules such as amino acids, proteins and nucleic acids and secondary metabolites. Similarly, photosynthesis requires large amounts of proteins which are involved in the photosynthetic process—Rubisco, the key enzyme for CO2 fixation, is the main N containing component of the cell—and a decrease in Rubisco activity has been shown to be accompanied by a decrease in N metabolism (Matt et al. 2002). During vegetative and early reproductive stages of wheat development, assimilated carbon is temporarily stored as carbohydrate, WSC and starch; these carbohydrate reserves are later remobilised and utilised.
Network analysis using the genes that were differentially expressed between the high and low WSC lines generated an interaction map that linked genes involved in starch and protein and amino acid metabolism via pyridoxal phosphate (PLP). PLP is an essential co-factor for many enzymes involved in amino acid metabolism; with the exception of glycogen phosphorylase, all enzymes that use PLP as a co-factor act upon amino acids (John 1995). PLP acts as a co-enzyme in all transamination reactions which is a critical step in the synthesis of some non-essential amino acids including glutamine and asparagine from aspartate and glutamate. Aspartate and alpha-ketoglutarate are synthesised by aspartate aminotransferase (AAT) from glutamate and oxaloacetate; α-ketoglutarate and oxaloacetate are key intermediates in the citric acid cycle. Glycogen phosphorylase (starch phosphorylase in plants) breaks down starch releasing glucose units in the form of glucose-1-phosphate which is subsequently converted to glucose-6-phosphate by phosphoglucomutase for metabolism including the citric acid cycle. PLP has been shown to increase the thermostability of AAT (De la Torre et al. 2007) and to be a strong inhibitor of glutamate dehydrogenase which catalyses the synthesis of glutamate from ammonia and α-ketoglutarate. In addition, PLP inhibits several glycolytic enzymes, including starch synthase, glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase (Teixeira and Davies 1974; Gao et al. 2004). Levels of PLP have been shown to be influenced by N status—PLP concentration is high under low N conditions and falls when N levels increase (Teixeira and Davies 1974). Therefore, based on this theory, high nitrogen favours ammonia incorporation into amino acids and promotes glycolysis, which leads to high biomass nitrogen and low WSC accumulation.
In this network study, expression of the two starch synthesis enzymes was higher in the high WSC lines compared to the low WSC lines, as was expression of GS and AAT, while expression of SHMT and MS were lower in the high WSC lines. These observations suggest that there was an increased flow of N to glutamine and oxaloacetate in the high WSC lines compared to the low WSC lines and less aspartate and methionine; methionine is synthesised from aspartate. Similarly, there was an increase in the synthesis of starch in the high WSC lines and a decrease in the amount of glycine/serine; serine is synthesised from 3-phosphoglycerate, an intermediate in the glycolysis and Calvin cycles.
Thaliana is the only plant with information in the STITCH database, necessitating the identification of Arabidopsis homologues of the differentially expressed wheat genes. Homologues were found for only 65% of the wheat genes. The use of the complete set of differentially expressed wheat genes as input into the STITCH 2.0 software may have yielded a more complete picture of the interactions between the C and N metabolic pathways.
Gene expression in response to environmental variation
Plant growth and development are highly dependent on the coordinated interaction between N and C metabolism. A complex multi-layer system interacts to balance the plant’s capacity to assimilate nitrogen with N availability, C metabolism—including photosynthesis, photorespiration and respiration—and its needs for growth and development. Little is known about the response of these genes in wheat or of the effect of environmental variation on expression of genes involved in the accumulation of stem carbohydrates in field-grown wheat plants.
As water and nitrogen are two of the most important environmental factors influencing levels of WSC in the field, we evaluated their effect on the levels of expression of 12 genes that were known to be differentially transcribed in high and low WSC lines. The 12 genes were selected as they represented different metabolic gene categories. In particular, they represented metabolic categories that were strongly differentially represented in the high and low WSC SB progeny, namely carbohydrate, cell wall or nitrogen/protein metabolism.
Of the 12 genes selected, only two genes, TaNIR1 and TaGd, both of which are involved in N metabolism, had a consistent relationship between their expression levels and stem WSC concentrations in high and low WSC lines and across lines in response to varying N and water levels. TaNIE1 encodes NIR1, a nitrate reductase that reduces nitrate to nitrite, which is subsequently converted to ammonium for synthesis of amino acids. TaGd encodes glycine decarboxylase complex H-protein (Gd), which is an important component of the glycine decarboxylase multi-enzymatic complex and the biosynthesis and activity of this multi-enzyme complex in plants appear to be determined by the biosynthesis of the H-protein (Bauwe and Kolukisaoglu 2003). Together with SHMT, Gd is involved in the reversible conversion of glycine to serine; SHMT also uses PLP as a co-factor. Interestingly, the expression levels of both Gd and SHMT were negatively associated with genotypic variation in stem WSCc (Supplementary Table 2). The Gd reaction also leads to production of methylated tetrahydrofolate which is an active one carbon unit for a number of important biosynthetic processes including synthesis of methionine, pyrimidines and purines. Indeed, as noted earlier, the expression levels of MS and S-adenosylmethionine synthetase also had an inverse relationship with stem WSCc between the high and low WSC lines in the transcript expression dataset (Supplementary Table 2). We observed that TaNIR1 and TaGd were expressed more strongly in low WSC lines and under high N and irrigated conditions. These observations are consistent with the higher biomass nitrogen percentage observed in the low WSC lines, and in the high N and irrigated managed environments, and in the higher percent grain protein in high N conditions.
Coordinated interaction between C and N metabolism in field-grown wheat plants
We have undertaken a global expression analysis using field-grown material from the phenotypic tails of a wheat population segregating for WSC to investigate the molecular mechanisms that control levels of WSC in the stems of field-grown wheat. The data presented in this study confirm the close interaction between C and N metabolism with different C and N metabolism-related gene categories differentially expressed between high and low WSC lines. High WSC lines had increased levels of expression of genes involved in starch synthesis and the citric acid cycle and a lower level of expression of genes involved in cell wall metabolism and amino acid synthesis, than low WSC lines. Using the set of differentially expressed genes as input, chemical–protein network analyses demonstrated a linkage between starch and N metabolism via the co-factor pyridoxal phosphate. Expression analysis of 12 C and N metabolism-related genes in high and low WSC lines sampled under varying environmental conditions (nitrogen level and soil water status) that affect WSC accumulation revealed that only the two nitrogen/amino acid metabolism genes remained consistent with the observed levels of WSC. Together these results indicate that high WSC lines are storing more carbon, taking up less nitrogen and synthesising less amino acids. The results suggest that the assimilation of nitrogen into amino acids controls the levels of WSC in the stems of field-grown wheat. Further research investigating expression and activity of more genes and metabolites involved in N and C metabolism under a broader range of environmental conditions that alter N and C metabolism are required to confirm these suggestions.
References
Bauwe H, Kolukisaoglu U (2003) Genetic manipulation of glycine decarboxylation. J Exp Bot 54:1523–1535
Bhuiyan NH, Liu WP, Liu GS, Selvaraj G, Wei YD, King J (2007) Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol Biol 64:305–318
Chalmers J, Lidgett A, Cummings N, Yingying C, Forster J, Spangenberg G (2005) Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol J 3:459–474
De la Torre F, Suarez MF, de Santis L, Canovas FM (2007) The aspartate aminotransferase family in conifers: biochemical analysis of a prokaryotic-type enzyme from maritime pine. Tree Phys 27:1283–1291
Dreccer MF, van Herwaarden AF, Chapman S (2009) Grain number and grain weight in lines contrasting for stem soluble carbohydrate concentration. Field Crops Research 112:43–54
Duggan BL, Richards RA, van Herwaarden AF (2005) Agronomic evaluation of a tiller inhibition gene (tin) in wheat. II. Growth and partitioning of assimilate. Aust J Agric Res 56:179–186
Foulkes MJ, Scott RK, Sylvester-Bradley R (2002) The ability of wheat cultivars to withstand drought in UK conditions: formation of grain yield. Journal of Agricultural Science 138:153–169
Gao Z, Keeling P, Shibles R, Guan HP (2004) Involvement of lysine-193 of the conserved “K-T-G-G” motif in the catalysis of maize starch synthase IIa. Archives of Biochem Biophys 427:1–7
Gebbing T, Schnyder H, Kuhbauch W (1999) The utilization of pre-anthesis reserves in grain filling of wheat. Assessment by steady-state (CO2)-C-13/(CO2)-C-12 labelling. Plant Cell Environ 22:851–858
González-Verdejo CI, Die JV, Nadal S, Di Pietro A, Barandiaran X, Cubero JI, Román B (2008) Isolation and expression analysis of a cobalamin-independent methionine synthase gene from the parasitic plant Orobanche ramosa. Scientia Horticulturae 116:337–341
Gutierrez RA, Lejay L, Dean A, Chiaromonte F, Shash DE, Coruzzi GM (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8:R7
John RA (1995) Pyridoxal phosphate-dependent enzymes. Biochem Biophys Acta 1248:81–96
Judel GK, Mengel K (1982) Effect of shading on non-structural carbohydrates and their turnover in culms and leaves during the grain filling period of spring wheat. Crop Sci 22:958–962
Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, Beyer A, Bork P (2010) STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res 38(suppl 1):D552–D556
Matt P, Krapp A, Haake V, Mock HP, Stitt M (2002) Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J 30:663–677
McGrath VB, Blakeney AB, Batten GD (1997) Fructan to nitrogen ratio as an indicator of nutrient stress in what crops. New Phytol 136:145–152
Nunes-Nesi A, Fernie AR, Stitt M (2010) Metabolic signalling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3:973–996
Olivares-Villegas JJ, Reynolds MP, McDonald GK (2007) Drought-adaptive attributes in the Seri/Babax hexaploid wheat population. Funct Plant Biol 34:189–203
R Development Core Team (2006) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rattey A, Shorter R (2010) Evaluation of CIMMYT conventional and synthetic spring wheat germplasm in rainfed sub-tropical environments. I. Grain yield. Field Crop Research 118:273–281
Rattey A, Shorter R, Chapman S, Dreccer F, van Herwaarden A (2009) Variation for and relationships among biomass and grain yield component traits conferring improved yield and grain weight in an elite wheat population grown in variable yield environments. Crop & Pasture Science 60:717–729
Rebetzke GJ, van Herwaarden AF, Jenkins C, Weiss M, Lewis D, Ruuska S, Tabe L, Fettell NA, Richards RA (2008) Quantitative trait loci for water-soluble carbohydrates and associations with agronomic traits in wheat. Aust J Agric Res 59:891–905
Ruuska SA, Rebetzke GJ, van Herwaarden AF, Richards RA, Fettell NA, Tabe L, Jenkins CLD (2006) Genotypic variation in water-soluble carbohydrate accumulation in wheat. Funct Plant Biol 33:799–809
Ruuska SA, Lewis DC, Kennedy G, Furbank RT, Jenkins CLD, Tabe LM (2008) Large scale transcriptome analysis of the effects of nitrogen nutrition on accumulation of stem carbohydrate reserves in reproductive stage wheat. Plant Mol Biol 66:15–32
Schnyder H (1993) The role of carbohydrate storage and redistribution in the source-sink relations of wheat and barley during grain filling—a review. New Phytol 123:233–245
Scofield GN, Ruuska SA, Aoki N, Lewis DC, Tabe LM, Jenkins CD (2009) Starch storage in the stems of wheat plants: localisation and temporal changes. Ann Bot 103:859–868
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Teixeira AR, Davies DD (1974) The control of plant glutamate dehydrogenase by pyridoxal-5′-phosphate. Phytochemistry 13:2071–2079
Thimm O, Blaesing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37(6):914–939
Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138(3):1195–1204
Van den Ende W, Clerens S, Vergauwen R, Van Riet L, Van Laere A, Yoshida M, Kawakami A (2003) Fructan 1-exohydrolases. β-(2,1)-trimmers during graminan biosynthesis in stems of wheat? Purification, characterisation, mass mapping and cloning of two fructan 1-exohydrolase isoforms. Plant Physiol 131:621–631
van Herwaarden AF, Angus JF, Richards RA, Farquhar GD (1998a) ‘Haying-off’, the negative grain yield response of dryland wheat to nitrogen fertiliser II. Carbohydrate and protein dynamics. Aust J Agric Res 49:1083–1093
van Herwaarden AF, Richards RA, Farquhar GD, Angus JF (1998b) ‘Haying’off’, the negative grainyield response of dryland wheat to nitrogen fertiliser III. The influence of water deficit and heat shock. Aust J Agric Res 49:1095–1110
van Herwaarden A, Richards R, Angus J (2003) Water-soluble carbohydrates and yield in wheat. Solutions for a better environment: Proceedings of the 11th Australian Agronomy Conference, Geelong, Victoria, Australia, 2–6 February 2003, 0–4
Wang C, van den Ende W, Tillberg JE (2000) Fructan accumulation induced by nitrogen deficiency in barley leaves correlates with the level of sucros:fructan 6-fructosyltransferase mRNA. Planta 211:701–707
Welham SJ, Gogel BJ, Smith AB, Thomson R, Cullis B (2006) Evaluation of models for late-stage variety evaluation trials. Australasian GenStat/StatGen Conference, Victor Harbour, Australia, p 44
Xue GP, McIntyre CL, Glassop D, Shorter R (2008a) Use of expression analysis to dissect alterations in carbohydrate metabolism in wheat leaves during drought stress. Plant Mol Biol 67:197–214
Xue G-P, McIntyre CL, Jenkins CLD, Glassop D, van Herwaarden AF, Shorter R (2008b) Molecular dissection of variation in carbohydrate metabolism related to water-soluble carbohydrate accumulation in stems of wheat. Plant Physiol 146:441–454
Yamada T, Matsuda F, Kasai K, Fukuoka S, Kitamura K, Tozawa Y, Miyagawa H, Wakasa K (2008) Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell 20:1316–1329
Yang J, Zhang J, Wang Z, Zhu Q, Liu L (2004) Activities of fructan- and sucrose- metabolizing enzymes in wheat stems subjected to water stress during grain filling. Planta 220:331–343
Yang DL, Jing RL, Chang XP, Li W (2007) Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 176:571–584
Zhang J, Huang S, Fosu-Nyarko J, Dell B, McNeil M, Waters I, Moolhuijzen P, Conocono E, Appels R (2009) The genome structure of the 1-FEH genes in wheat (Triticum aestivum L.): new markers to track stem carbohydrates and grain filling QTLs in breeding. Mol Breeding 22:339–351
Acknowledgements
We thank Axel Nagel for making a test version of the Mercator pipeline available before publication. The development of the pipeline was supported by the BMBF Project MAPMEN (0315049A). In addition, we thank Greg Roberts, Terry Collins, Philip van Drie, Laura Barnes and Janneke Drenth for their excellent technical assistance with aspects of these experiments. This research was jointly funded by CSIRO and the Grains Research and Development Corporation of Australia.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Interaction maps of A. thaliana homologues of wheat transcripts differentially regulated between individuals with high and low WSC with one or more input loci. White circles: input loci; pink circles: predicted interacting proteins (Supplementary Table 3); green squares: predicted interacting metabolites. Blue line: protein–protein interaction; green line: protein–metabolite interaction; red line: metabolite–metabolite interaction (PPT 599 kb)
Supplementary Table 1
Primers for real-time PCR analysis of Triticum aestivum genes (XLS 207 kb)
Supplementary Table 2
Probe sets that were differentially expressed between high and low WSC lines. (XLS 207 kb)
Supplementary Table 3
List of interactors generated by STITCH 2.0 interaction map analysis at a confidence level of 0.900 or less. (XLS 38 kb)
Rights and permissions
About this article
Cite this article
McIntyre, C.L., Casu, R.E., Rattey, A. et al. Linked gene networks involved in nitrogen and carbon metabolism and levels of water-soluble carbohydrate accumulation in wheat stems. Funct Integr Genomics 11, 585–597 (2011). https://doi.org/10.1007/s10142-011-0232-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0232-5