Abstract
The chemical 2,4-dichlorophenoxyacetic acid (2,4-D) regulates plant growth and development and mimics auxins in exhibiting a biphasic mode of action. Although gene regulation in response to the natural auxin indole acetic acid (IAA) has been examined, the molecular mode of action of 2,4-D is poorly understood. Data from biochemical studies, (Grossmann (2000) Mode of action of auxin herbicides: a new ending to a long, drawn out story. Trends Plant Sci 5:506–508) proposed that at high concentrations, auxins and auxinic herbicides induced the plant hormones ethylene and abscisic acid (ABA), leading to inhibited plant growth and senescence. Further, in a recent gene expression study (Raghavan et al. (2005) Effect of herbicidal application of 2,4-dichlorophenoxyacetic acid in Arabidopsis. Funct Integr Genomics 5:4–17), we have confirmed that at high concentrations, 2,4-D induced the expression of the gene NCED1, which encodes 9-cis-epoxycarotenoid dioxygenase, a key regulatory enzyme of ABA biosynthesis. To understand the concentration-dependent mode of action of 2,4-D, we further examined the regulation of whole genome of Arabidopsis in response to a range of 2,4-D concentrations from 0.001 to 1.0 mM, using the ATH1-121501 Arabidopsis whole genome microarray developed by Affymetrix. Results of this study indicated that 2,4-D induced the expression of auxin-response genes (IAA1, IAA13, IAA19) at both auxinic and herbicidal levels of application, whereas the TIR1 and ASK1 genes, which are associated with ubiquitin-mediated auxin signalling, were down-regulated in response to low concentrations of 2,4-D application. It was also observed that in response to low concentrations of 2,4-D, ethylene biosynthesis was induced, as suggested by the up-regulation of genes encoding 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase. Although genes involved in ethylene biosynthesis were not regulated in response to 0.1 and 1.0 mM 2,4-D, ethylene signalling was induced as indicated by the down-regulation of CTR1 and ERS, both of which play a key role in the ethylene signalling pathway. In response to 1.0 mM 2,4-D, both ABA biosynthesis and signalling were induced, in contrast to the response to lower concentrations of 2,4-D where ABA biosynthesis was suppressed. We present a comprehensive model indicating a molecular mode of action for 2,4-D in Arabidopsis and the effects of this growth regulator on the auxin, ethylene and abscisic acid pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The chemical 2,4-dichlorophenoxyacetic acid (2,4-D), a phenoxycarboxylic acid, regulates growth and development of plants. At low concentrations, it induces growth, whereas at high concentrations, it is herbicidal. In tissue culture media, 2,4-D is used at a concentration of 1–50×10−4 mM to induce growth (Schmidt and Willmitzer 1991). In agriculture, it is used at very high concentrations as a selective herbicide to destroy dicotyledonous weeds among monocotyledonous crops (Grabinska-Sota et al. 2003). The concentration-dependent biphasic mode of action of 2,4-D is similar to naturally occurring auxins such as indole acetic acid (IAA), and therefore, 2,4-D is classified as an auxinic herbicide (Grossmann 2000).
Auxins play a key role in a variety of growth and developmental processes in plants by regulating cell division, elongation and differentiation (Bennett et al. 1998; Becker and Hedrich 2002). Auxins have been shown to induce the expression of several genes within minutes of application (Abel and Theologis 1996). These genes, known as early-response genes, fall into three major categories: small auxin up-regulated genes (SAURs), the Aux/IAA genes and GH3s (Abel and Theologis 1996; Hagen and Guilfoyle 2002). The three gene families have been well characterised in Arabidopsis, while little is known in monocots. A recent study has shown that although members of the GH3 family are highly conserved in the dicots and monocots, a subgroup of the GH3 family that is present in Arabidopsis is clearly not found in rice (Jain et al. 2005).
The Aux/IAA genes are involved in structural and regulatory functions and encode short-lived proteins (Reed 2001). The turnover of these auxin-induced proteins known to occur via the ubiquitin-mediated auxin signalling is essential for auxin-induced response (Leyser 2002). The F-box proteins are involved in selecting the specific auxin-induced protein for degradation via the ubiquitin–proteosome pathway (del Pozo and Estelle 2000). The TIR1 gene, which encodes an F-box protein, has been identified to play a significant role in auxin signalling (Gray et al. 1999; Dharmasiri and Estelle 2002; Kepinski and Leyser 2005) and is an auxin receptor (Dharmasiri et al. 2005). In this study, we examined the regulation of genes associated with the auxin signalling pathway in response to 2,4-D applications.
Grossmann (2000) proposed that at high concentrations, both natural auxins and auxinic herbicides induced the plant hormone ethylene, which in turn triggered the biosynthesis of another plant hormone, abscisic acid (ABA). This model proposed that auxins at high concentrations increase the enzyme activity of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, the key regulatory enzyme in ethylene biosynthesis (Sato and Theologis 1989). The model was based on data derived from biochemical studies that measured ACC synthase activity and ethylene levels (Hansen and Grossmann 2000), whereas the current study examined the regulation of genes involved in ethylene biosynthesis and ethylene signalling pathway in response to 2,4-D application.
Hansen and Grossmann (2000) observed that it was the auxin-induced ethylene that triggered xanthophyll cleavage, leading to ABA biosynthesis. The key regulatory enzyme of ABA biosynthesis, 9-cis-epoxycarotenoid dioxygenase, is involved in xanthophyll cleavage, which in turn leads to the production of xanthoxin/xanthoxal (Cutler and Krochko 1999; Qin and Zeevaart 1999; Grossmann 2000). Studies using a tomato mutant (Never-ripe), known to be deficient in ethylene perception, support the proposal that ethylene, induced by high concentrations of auxins or auxinic herbicides, leads to increased ABA biosynthesis (Hansen and Grossmann 2000). In an earlier study (Raghavan et al. 2005), we reported that 2,4-D at herbicidal levels induced ABA biosynthesis, leading to ABA signalling and response. In order to further understand the molecular mode of action of 2,4-D in relation to the key plant hormone pathways and their interactions, we examine the regulation of genes associated with ABA and ethylene biosynthesis and signalling in response to lower concentrations of 2,4-D. Based on the gene expression results of this study, we present a comprehensive model of the molecular responses of Arabidopsis genes involved in auxin, ethylene and ABA pathways to varying concentrations of 2,4-D.
Results and discussion
Microarray data
Gene expression of each treatment was compared with the control, and a twofold change in expression and signal value greater than 100 was considered significant. Refer to Raghavan et al. (2005) for the procedure used to select significantly regulated genes.
Biological duplication of the 1.0-mM 2,4-D treatment confirmed the reliability of the microarray data (Raghavan et al. 2005). Comparison of array results from the biological duplicates of the 1.0 mM 2,4-D treatment indicated that 99.2% of the genes that were called present in both duplicates (13,480) showed a variation coefficient of less than 50%, indicating an extremely high degree of reproducibility between the biological replicates, and scatter plot analysis of the signal values from treatment duplicates further support this as 80% of probes on both arrays were within a twofold difference. Results of this analysis have been discussed in Raghavan et al. (2005). Data analysis showed that there was low biological variability and technical variability between the duplicates. Based on the reliability of using the Affymetrix platform, we have used data obtained from single chip hybridization. Further, the scaling factor of all arrays was below threefold, indicating that there was no significant variability in the assays and suggesting a high level of reproducibility (Table 1).
Semi-quantitative polymerase chain reaction
Semi-quantitative polymerase chain reaction (PCR) was used to verify the expression pattern of selected genes of interest. Semi-quantitative PCR has often been selected to verify microarray results due to its low cost and technical simplicity. Examining the PCR products on agarose gels indicated uniform expression of housekeeping genes encoding GAPDH (GapC) and actin under all conditions (control and 2,4-D treatments) (Fig. 1). All other genes (IAA2, IAA19, IAA13, AtERF4, ABA induced PP2C and ABF3) exhibited differential expression under the influence of different concentrations of 2,4-D (Fig. 1). The auxin-induced gene IAA2 was up-regulated at all concentrations compared with the control. Gene IAA19 was also up-regulated compared with the control, except in the case of the 0.001 mM 2,4-D treatment. Expression levels of the gene encoding auxin-regulated protein IAA13 indicated that it was expressed during all treatments, with a clear increase in expression at 0.1 and 1.0 mM 2,4-D. The expression of the gene encoding ethylene response factor (AtERF4) showed a high level of expression in the control and almost similar levels of expression at all treatments. The expression pattern of ABA-induced PP2C showed an increase in expression levels in response to 0.1 and 1.0 mM 2,4-D compared with the control and was not detected in response to 0.01 mM 2,4-D. The ABF3 gene was down-regulated at 0.01 mM 2,4-D and up-regulated at 1.0 mM 2,4-D in comparison with the control.
Examination of gene expression using semi-quantitative PCR. Changes in gene expression levels from microarray experiments of corresponding probe sets/genes compared with the control are shown in the table alongside. The genes selected are associated with the auxin/ethylene/abscisic acid pathways. Total RNA (5 μg) was reverse-transcribed, and PCRs were conducted for 25 cycles using equal quantities of 100× diluted cDNA as template. GAPDH and actin represented housekeeping genes. C control, Conc concentration
The six genes that were selected for verification using semi-quantitative PCR were generally consistent with the microarray results (Fig. 1). For example, the IAA2 gene showed an increase in transcript levels at the 0.001, 0.01 and 0.1 mM 2,4-D treatments compared with the control. Another example was the ABF3 gene, which showed a significant decrease in transcript levels at the 0.1-mM 2,4-D treatment and an increase at the 1.0-mM 2,4-D treatment compared with the control. However, in spite of repeated PCR, inconsistency was observed in transcript levels for the IAA19 gene at 0.001 mM. The increase in transcript level indicated by the microarray results was not observed in PCR experiments.
Regulation of auxin response and signalling by 2,4-D
The plant growth regulant 2,4-D up-regulated gene expression of a large number of auxin response genes, irrespective of the concentration of 2,4-D (Table 2). In response to all concentrations of 2,4-D tested in this study, the genes IAA1, IAA13 and IAA19 were all up-regulated (Table 2), confirming the auxin-like property of 2,4-D. Also HAT2, a gene encoding a member of the HD-ZIP family of transcription factors, was induced in response to all concentrations of 2,4-D tested. Other gene expression studies have identified HAT2 as an auxin-inducible gene (Sawa et al. 2002), which is consistent with the observations in the present study.
The degradation of auxin-induced proteins via the 26S proteosome is essential for proper auxin response in plants (Leyser 2002). Auxin-induced growth and development appears to be controlled by ubiquitin-mediated auxin signalling (Leyser 2001). Gene expression levels indicated a decrease in expression levels of ASK1 and TIR1 in response to 0.001 and 0.01 mM 2,4-D application (Table 2). Both TIR1 and ASK1 encode proteins of the SCF complex (Kepinski and Leyser 2002), which play a key role in the selection of Aux/IAA proteins for degradation via the 26S proteosome (Dharmasiri and Estelle 2002). The interaction of TIR1 and ASK1 is essential for auxin signalling and thereby the auxin response in plants (Gray et al. 1999). Competitive binding experiments have shown that auxins exhibit affinity to TIR1 and further indicated that IAA has a greater affinity compared with 2,4-D or naphthalene acetic acid (NAA) (Dharmasiri et al. 2005). This may suggest that at 0.001 and 0.1 mM, 2,4-D induced affected auxin response and signalling.
Mutants of TIR1 and ASK1 exhibit reduced lateral root formation (Gray et al. 1999; Kepinski and Leyser 2002); consistent with this is the observation that plants exposed to a range of 2,4-D concentrations (0.001, 0.01, 0.1 and 1.0 mM) exhibited a clear inhibition of lateral root development (Fig. 2). Interestingly, in response to both 0.1 and 1.0 mM 2,4-D, there was an increase in expression of genes encoding auxin-induced proteins; however, there was no significant regulation of either TIR1 or ASK1 (Table 2).
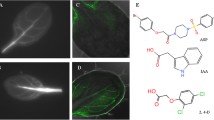
Lateral root development. Photos of Arabidopsis plants under control conditions and 48 h post treatment with 2,4-D. Control plant exhibits profuse lateral root formation, while in treated plants, lateral root development was inhibited. Plants treated with 2,4-D show epinastic growth-elongated petioles and leaf curling. In response to 0.1 and 1.0 mM 2,4-D, the leaves turn brown and exhibit senescence
The plant response to 0.01 mM 2,4-D appears to be complex, as the results indicate up-regulation of AUX1, GH3, AIR12 and other genes encoding auxin-regulated proteins and down-regulation of SAUR, IAA17 and another auxin-induced protein (Table 2). The AUX1 gene encodes a permease involved in hormone transport and lateral root formation (Marchant et al. 2002), and the recessive mutant aux1 in Arabidopsis confers resistance to auxin (Timpte et al. 1995). Studies have identified AUX1 as a target site for 2,4-D, with point mutations in this gene conferring herbicide resistance in Arabidopsis (Roux et al. 2004). The increase in expression of AUX1 may imply the involvement of this gene product in the transport of 2,4-D, and inhibited lateral root formation may be due to the down-regulation of TIR1. Many auxin-induced proteins have been shown to inhibit auxin-response, while IAA17 has been shown to induce an auxin response (Reed 2001). We observed the down-regulation of IAA17 and an up-regulation of IAA1 in response to 0.01 mM 2,4-D (Table 2), suggesting that 2,4-D inhibited an auxin-response.
Analysis of gene expression indicated that 2,4-D suppressed the auxin response at all concentrations tested in this study. We further observed that in response to low concentrations of 2,4-D, auxin signalling was repressed, whereas, at higher concentrations, there was no significant regulation of genes involved in auxin signalling. The epinastic growth and inhibition of lateral root development in response to 2,4-D may be attributed to improper auxin signalling (Figs. 2 and 3). The concentration dependent regulation of TIR1 and ASK1 may suggest their involvement in the biphasic mode of action of 2,4-D. Further evaluation of the effect of 2,4-D on Arabidopsis lines mutated in the genes for TIR1 and ASK1 would validate their functions in regulating the mode of action of 2,4-D.
2,4-D regulated genes involved in ethylene biosynthesis ethylene signal transduction pathways
The model for the mode of action of high concentration of auxins and auxinic herbicides proposed by Grossmann (2000) was based on biochemical studies measuring the activity of key enzymes and the concentration of metabolic products (Hansen and Grossmann 2000), while the results discussed here are based on the transcriptome analysis.
The amino acid methionine is the precursor of ethylene, with the first step in the pathway of ethylene biosynthesis being the conversion of methionine to S-adenosylmethionine (SAM/AdoMet) by the enzyme SAM synthetase (Schaller and Kieber 2002). The SAM is then converted to ACC by ACC synthase, the rate-limiting enzyme of ethylene biosynthesis (Sato and Theologis 1989), with the final step being the conversion of ACC to ethylene in the presence of ACC oxidase (Zarembinski and Theologis 1994; Schaller and Kieber 2002; Wang et al. 2002). The final enzymatic step, the conversion of ACC to ethylene is accompanied by cyanide formation (Grossmann 1996).
Transcriptome analysis indicates that there was no change in transcript level for any of the genes encoding enzymes involved in ethylene biosynthesis for either of the higher concentrations (0.1 or 1.0 mM) of 2,4-D applied to the roots of Arabidopsis, and yet there was regulation of genes involved in the ethylene signalling pathway (Table 3). The transcriptional response to 0.001 mM 2,4-D, the lowest concentration of 2,4-D used in this study, indicated an increase in transcript of gene encoding ACC oxidase (At5g63600). However, functional analysis using the annotations from The Arabidopsis Information Resources (TAIR) database (April 2005) indicated that the At5g63600 gene was involved in ferredoxin hydrogenase activity and as such may not indicate changes directly in the ethylene biosynthesis.
Gene expression analysis of the response to 0.01 mM 2,4-D indicated a significant decrease in the transcript levels for SAM synthetase (Table 3), but concurrently there was an increase in transcript levels for gene AtACS6, a gene encoding one member of the ACC synthase family. The Arabidopsis ACC synthase multigene family is characterised by members showing differential expression during development and response to environmental stimuli (Liang et al. 1992). The gene AtACS6 has been identified as being multi-responsive (Arteca and Arteca 1999), and thereby, the increased transcription of AtACS6 may be in response to stress induced by 2,4-D application. It is interesting to note that under the conditions used in this study, AtACS6 was the only member of the multigene family encoding ACC synthase that was regulated by 2,4-D.
In Arabidopsis, a protein family of five members (ETR1, ETR2, ERS1, ERS2 and EIN4) has been identified as comprising ethylene receptors (Hua et al. 1998; Hall et al. 2000; Schaller and Kieber 2002). These receptors share homology with the two-component histidine kinases of bacteria (Wang et al. 2003). In Arabidopsis, these receptors are localised in the endoplasmic reticulum and are involved in ethylene perception (Gao et al. 2003; Alonso and Stepanova 2004). The receptors play a key role in regulating downstream signalling of the hormone (Chang and Shockey 1999; Schaller and Kieber 2002; Alonso and Stepanova 2004). Ethylene receptors are negative regulators of ethylene signalling (Schaller and Kieber 2002). Results indicated the down-regulation of ERS only in response to 1.0 mM 2,4-D, suggesting that ethylene signalling was induced even if there was no regulation of ethylene biosynthesis genes in response to 1.0 mM 2,4-D. Although the results suggested induced ethylene biosynthesis in response to 0.001 and 0.01 mM 2,4-D application, there was no significant regulation of genes encoding ethylene receptors at these levels of 2,4-D (Fig. 3).
A model representing the molecular mode of action of varying concentrations of 2,4-D on a auxin, b ethylene and c abscisic acid pathways. Dotted boxes indicate involvement in biosynthesis. Red font indicates up-regulation and green font indicates down-regulation. Dotted lines indicate a possible mode of action. Refer to Tables 2, 3, and 4 for details of the genes
In Arabidopsis, ethylene is known to cause radial swelling of dark grown hypocotyls, and it also inhibits growth of the root and hypocotyl, with these responses collectively referred to as the “triple response” (Guo and Ecker 2004). Screening has identified the mutant crt1 in Arabidopsis, which exhibits a constitutive triple response (CTR) in the absence of ethylene (Schaller and Kieber 2002; Guo and Ecker 2004). Like the ethylene receptors, the CTR1 protein is localised in the endoplasmic reticulum (Gao et al. 2003) and shows similarity to the mitogen-activated protein kinase kinase kinase (MAPKKKs) (Chang 2003; Ouaked et al. 2003). Genetic studies (Huang et al. 2003) confirmed that the CTR1 gene is a negative regulator of ethylene signalling. In the absence of ethylene, the receptors repress the ethylene-related responses via activation of CTR1 (Chang and Shockey 1999). Results from our study indicated down-regulation of CTR1 in response to 0.1 and 1.0 mM 2,4-D (Table 3), suggesting that application of these levels of 2,4-D induced an ethylene response.
Studies have shown that the kinase domain of ETR1 and ERS1 (receptors of ethylene) can interact with CTR1 (Wang et al. 2002) to switch off the ethylene-signalling pathway (Huang et al. 2003). Consistent with their regulatory roles in modulating the ethylene response (disruption of the interaction between CTR1 and ETR1), the presence of ethylene results in the induction of ethylene signalling (Guo and Ecker 2004). The down-regulation of CTR1 is suggestive of positive regulation of ethylene signalling pathway by 0.1 and 1.0 mM 2,4-D. It was interesting to observe that although there was no significant regulation of genes involved in ethylene biosynthesis in response to 0.1 and 1.0 mM 2,4-D, genes encoding ethylene receptors and CTR1 were down-regulated, indicating that at high concentrations of 2,4-D, the ethylene response however was induced (Fig. 3).
In the ethylene signal transduction pathway downstream to CTR1 is EIN2, which consists of an integral membrane domain (Chang and Shockey 1999). The EIN2 gene is a positive regulator of ethylene signalling (Alonso and Stepanova 2004); however, results of this study did not identify regulation of EIN2 by 2,4-D. Downstream of EIN2 are two types of transcription factors, EIN3/EIL (Guo and Ecker 2004), with EIN3 a positive regulator of ethylene signalling (Schaller and Kieber 2002). EIN3 and EIL bind to the promoter region of the ethylene response factor (ERF1) gene (Schaller and Kieber 2002). ERF1, an EREBP transcription factor (Fujimoto et al. 2000), is induced by EIN3/EIL, indicating the sequential involvement of transcription factors in ethylene signal transduction (Riechmann and Meyerowitz 1998; Schaller and Kieber 2002; Alonso and Stepanova 2004). Interestingly, the results indicated down-regulation of EIL1 in response to 0.001 mM 2,4-D application (Table 3), while there was no regulation of the gene encoding EIN3 or ERF1. This suggests that down-stream of CTR1, the ethylene-dependent signalling pathway was disrupted by 2,4-D at all concentrations tested, since there was no significant regulation of key positive regulators of ethylene signalling.
Gene expression results indicated the regulation of ERF transcription factors in response to 2,4-D application (Table 3). About 11 members of ERFs have been identified from the Arabidopsis genome (Schaller and Kieber 2002). Gene expression studies have further indicated the regulation of genes involved in ethylene signalling in response to various environmental stresses (Chen et al. 2002b). The regulation of ERFs may occur in an ethylene-dependent or ethylene-independent manner by biotic and abiotic stress (Fujimoto et al. 2000). Members of this family have a conserved DNA binding domain that displays specific binding to the GCC-box present in many stress-response genes (Fujimoto et al. 2000). The AtERF1, AtERF2 and AtERF5 genes are activators, while AtERF3 and AtERF4 genes are repressors of GCC box-dependent transcription (Fujimoto et al. 2000).
Gene expression results indicated up-regulation of AtERF4 in response to all concentrations of 2,4-D tested in this study and up-regulation of AtERF6 in response to 0.001 mM 2,4-D application (Table 3). Studies have shown that ERFs maybe regulated in an ethylene-independent manner in response to abiotic and biotic stress, and therefore, AtERF4 and AtERF6 may be regulated in an independent manner by 2,4-D (Fig. 3). A time-series experiment within the 1-h time period, to examine the gene regulation in response to 2,4-D, may explain a series of events leading to ethylene response.
2,4-D induced regulation of genes involved in ABA biosynthesis and signalling
The phytohormone ABA is a cleavage product of major epoxycarotenoids. The oxidative cleavage of 9-cis-neoxanthin and 9-cis-violaxanthin (9-cis-epoxycarotenoids) by 9-cis-epoxycarotenoid dioxygenase (NCED) (Cutler and Krochko 1999; Qin and Zeevaart 1999) results in the formation of xanthoxin/xanthoxal (Taylor et al. 2000). The carotenoid neoxanthin is formed from violaxanthin, a product of the xanthophyll cycle. Hansen and Grossmann (2000) observed no change in levels of neoxanthin, violaxanthin and antheraxanthin in response to auxins in cleavers and confirmed that the increase in levels of xanthoxal was an indirect influence of the auxin.
Whole genome expression results indicated that ABA biosynthesis was suppressed at low concentrations of 2,4-D, whereas at higher concentrations, ABA biosynthesis was induced significantly. In response to 0.01 mM 2,4-D, there was a decrease in the transcript level for the gene encoding zeaxanthin epoxidase (Table 4). Zeaxanthin epoxidase is involved in the conversion of zeaxanthin to violaxanthin via antheraxanthin in the xanthophylls cycle (Audran et al. 1998). Further, the TAIR Gene Ontology (GO) database annotates this gene to be associated with ABA biosynthesis. In response to 0.1 mM 2,4-D, there was a significant decrease in expression of the gene LOX2, which has also been associated with ABA biosynthesis (TAIR).
We have earlier reported a significant up-regulation of gene (NCED) encoding 9-cis-epoxycarotenoid dioxygenase only in response to 1.0 mM 2,4-D application (Raghavan et al. 2005), and now we show that this response was specific only to 1.0 mM 2,4-D and not lower concentrations of 2,4-D application. Furthermore, there was also significant increase in expression of the gene encoding delta-1-pyroline-5-carboxylate synthetase, which is also associated with ABA biosynthesis (Hoth et al. 2002), therefore suggesting the involvement of ABA in 2,4-D-induced senescence.
G proteins, protein phosphatases and protein kinases are all shown to be ABA-response regulators (Himmelbach et al. 2003). The genes ABI1 and ABI2 encode the serine/threonine protein phosphatases type 2C (PP2C), which are involved in ABA signalling (Meyer et al. 1994; Leung et al. 1997), and Arabidopsis mutants abi1-1 and aba2-1 indicated decreased ABA sensitivity (Himmelbach et al. 2003). ABI1 and ABI2 are both negative regulators of the ABA-response (Himmelbach et al. 2003). Gene expression results in this study indicated a significant increase in ABI1 in response to all concentrations of 2,4-D tested (Table 4). Phosphatase activity of ABI1 and ABI2, which are induced by ABA, de-phosphorylates the ABA signalling element, making it inactive (Merlot et al. 2001). Furthermore, in response to all concentrations of 2,4-D tested, there was an increase in expression of HVA22. HVA22 is induced both by ABA and stress conditions (Chen et al. 2002a). Since in response to lower concentrations of 2,4-D, there was no regulation of genes involved in ABA biosynthesis, the regulation of HVA22 may be due to induced stress. Further, the results also indicated the up-regulation of COR78, COR414, Kin1 and TAG1 (Table 4), which are induced in response to ABA and other stress responses (TAIR).
ABA induces the activity of SNF1-related protein kinase (Himmelbach et al. 2003), which is involved in hormonal and stress responses (Farras et al. 2001). The SNF1-related protein kinases, phosphorylate and activate the ABA-response element (ABRE) binding factor (ABF) (Himmelbach et al. 2003). The ABFs belong to basic domain leucine zipper (bZIP) family of transcription factors (Choi et al. 2000) and are induced by ABA and are differentially regulated by various environmental stresses (Choi et al. 2000). Gene expression results indicated significant up-regulation of ABF3 only in response to 1.0 mM 2,4-D, suggesting induced ABA signalling and response. The gene AtHB12, which codes for a homeobox-leucine zipper, was up-regulated in response to 0.1 and 1.0 mM 2,4-D (Table 4). AtHB12 is induced by ABA (Hoth et al. 2002) and osmotic stress (TAIR). Further, there was a significant up-regulation of RD20 in response to 1.0 mM 2,4-D. RD20 is induced by ABA, salinity and drought (Takahashi et al. 2000; Hoth et al. 2002).
Conclusions
Given that there is significant overlap in the set of genes induced by ABA and the genes induced by other stress responses, further studies need to be conducted to confirm if the regulation of genes observed in Arabidopsis in response to 2,4-D was a direct result of stress-induced ABA or by a more general stress response in the plant (Fig. 3). What is clear, however, is that herbicidal levels 2,4-D induce stress and senescence that are accompanied by ABA biosynthesis and signalling (Raghavan et al. 2005). The gene regulation observed in this present study is due to 2,4-D, but the exact molecular basis for this response remains unclear. Whilst a large number of transcription factors were regulated in response to 2,4-D, very few of these genes have been assigned to specific pathways. However, using the regulation of genes that can be assigned to specific roles in signal transduction pathways and plant metabolism, this study has developed a number of schemes that provide a mechanism to describe the molecular mode of action in Arabidopsis under the influence of varying concentrations of 2,4-D and, at the same time, attempts to identify those genes regulated by a more general stress response in the plant. As such, this study has identified genes that hold a key to further explain the molecular mode of action of this plant hormone herbicide. Further reverse genetics studies with mutant lines and specific genetic inhibition or gene knockout studies will help clarify the functional role individual genes play in the different plant responses to 2,4-D.
Materials and methods
Plant material and treatment
Surface sterilised seeds of Arabidopsis thaliana ecotype Columbia were grown on 30-ml half-strength Murashige and Skoog medium supplemented with 1.5 (w/v) sucrose and vitamin and solidified with 1% (w/v) phytagel in deep Petri dishes. Twelve seeds per plate were sown. Plates were kept in the dark at 4°C for 48 h, after which they were grown in culture rooms at 22°C and a 16-h photoperiod.
Plants were treated with 2,4-D 14 days after sowing. The plants developed four rosette leaves measuring greater than 1 mm and represented the 1.04 stage described by Boyes et al. (2001). Plants (14 days old) were treated with 2,4-D (BDH Chemical) for a period of 1 h. The chemical 2,4-D was dissolved in water, and the pH was adjusted to 7.0. The concentrations of 2,4-D tested in this study were 0.001, 0.01, 0.1 and 1.0 mM. Plates were flooded with 1 ml of stock solutions of 2,4-D. Control plants were flooded with 1 ml distilled water.
Target preparation and hybridization for microarray experiments
The whole genome oligonucleotide microarray, ATH1-121501, developed by Affymetrix was used in this study as the probe. Total RNA was extracted from 100 mg of plant material and used to generate the target for hybridization. Biological duplicates of the 1.0-mM 2,4-D treatment were used to check the reproducibility of microarray experiments. For biological duplication, two sets of plants were separately treated and RNA extracted from each were separately used to generate cRNA. Results from both the hybridisations were used to verify the reliability of expression results. For additional details of the procedure, refer to Raghavan et al. (2005).
Data analysis
Microarray data analysis was conducted using Microarray Suite version 5 (MAS5.0 developed by Affymetrix). The MAS 5.0 software was used for image, absolute and comparison analysis. The images of the hybridized arrays were checked for artefacts, followed by absolute analysis. Absolute analysis generated a signal value and a detection call for all probe sets. Signal values were generated based on the one-step Tukey’s biweight estimate, and detection calls were made based on Wilcoxon’s signed rank test (Affymetrix 2002b). The parameters used for microarray data analysis are given in Table 5.
Comparison analysis was conducted using the MAS 5.0 (Affymetrix 2002a). Data from each 2,4-D treatment (0.001, 0.01, 0.1 and 1.0 mM; array 1; array 2) was compared with the control, which served as the baseline data. All probes on the array were used for comparison analysis. The intensity of probes from the experiment and the baseline was scaled to a target value (TGT)=50. Furthermore, the MAS 5.0 software conducts a ‘robust normalisation’, which accounts for differences due to the affinity and linearity of probe sequences.
Comparison analysis uses the Wilcoxon’s signed-rank test to compute the change p value that indicates the likelihood of the significant change (Affymetrix 2002b). The default value for the tuneable parameter ‘gamma’ was used to assign a change call. The one-step Tukey’s biweight method was used to calculate the signal log ratio, which indicates the extent of change or fold change. The log base of scale 2 was used, and therefore, a signal log ratio of 1.0 indicated an increase by twofold and a signal log ratio of -1.0 indicated a decrease by twofold (Affymetrix 2002a,b).
Data were transferred for data mining to the DMT (Data Mining Tool). The DMT was used for selecting genes that were significantly up- and down-regulated (Affymetrix Data Mining Tool User’s Guide). Gene annotations were obtained from the GO database of the TAIR. Genes exhibiting significant regulation under the influence of varying concentrations of 2, 4-D can be found in the supplementary data files.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- ABA:
-
abscisic acid
- IAA:
-
indole acetic acid
- NAA:
-
naphthalene acetic acid
- TAIR:
-
The Arabidopsis Information Resources
References
Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111:9–17
Affymetrix (2002a) GeneChip expression analysis. Technical note
Affymetrix (2002b) Statistical algorithms description document. Technical note
Alonso JM, Stepanova AN (2004) The ethylene signaling pathway. Science 306:1513–1515
Arteca JM, Arteca RN (1999) A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature Arabidopsis leaves. Plant Mol Biol 39:209–219
Audran C, Borel C, Frey A, Sotta B, Meyer C, Simonneau T, Marion-Poll A (1998) Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol 118:1021–1028
Becker D, Hedrich R (2002) Channelling auxin action: modulation of ion transport by indole-3-acetic acid. Plant Mol Biol 49:349–356
Bennett MJ, Marchant A, May ST, Swarup R (1998) Going the distance with auxin: unravelling the molecular basis of auxin transport. Philos Trans R Soc Lond B Biol Sci 353:1511–1515
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510
Chang C (2003) Ethylene signalling: the MAPK module has finally landed. Trends Plant Sci 8:365–368
Chang C, Shockey JA (1999) The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol 2:352–358
Chen CN, Chu CC, Zentella R, Pan SM, Ho TH (2002a) AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol Biol 49:633–644
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T (2002b) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Cutler AJ, Krochko JE (1999) Formation and breakdown of ABA. Trends Plant Sci 4:472–478
del Pozo JC, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44:123–128
Dharmasiri S, Estelle M (2002) The role of regulated protein degradation in auxin response. Plant Mol Biol 49:401–409
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. Embo J 20:2742–2756
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12:393–404
Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signalling complexes. J Biol Chem 278:34725–34732
Grabinska-Sota EE, Wisniowska E, Kalka J (2003) Toxicity of selected synthetic auxines-2,4-D and MCPA derivatives to broad-leaved and cereal plants. Crop Prot 22:355–360
Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13:1678–1691
Grossmann K (1996) A role for cyanide, derived from ethylene biosynthesis, in the development of stress symptoms. Physiol Plant 97:772–775
Grossmann K (2000) Mode of action of auxin herbicides: a new ending to a long, drawn out story. Trends Plant Sci 5:506–508
Guo H, Ecker JR (2004) The ethylene signalling pathway: new insights. Curr Opin Plant Biol 7:40–49
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB (2000) Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol 123:1449–1458
Hansen H, Grossmann K (2000) Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol 124:1437–1448
Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signalling. Curr Opin Plant Biol 6:470–479
Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115:4891–4900
Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10:1321–1332
Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signalling in Arabidopsis. Plant J 33:221–233
Jain M, Kaur N, Tyagi AK, Khurana JP (2005) The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct Integr Genomics (online first). DOI 10.1007/s10142-005-0142-5
Kepinski S, Leyser O (2002) Ubiquitination and auxin signalling: a degrading story. Plant Cell 14(Suppl):S81–S95
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451
Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9:759–771
Leyser O (2001) Auxin signalling: the beginning, the middle and the end. Curr Opin Plant Biol 4:382–386
Leyser O (2002) Molecular genetics of auxin signalling. Annu Rev Plant Biol 53:377–398
Liang X, Abel S, Keller JA, Shen NF, Theologis A (1992) The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci U S A 89:11046–11050
Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14:589–597
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303
Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264:1452–1455
Ouaked F, Rozhon W, Lecourieux D, Hirt H (2003) A MAPK pathway mediates ethylene signalling in plants. Embo J 22:1282–1288
Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci U S A 96:15354–15361
Raghavan C, Ong EK, Dalling MJ, Stevenson TW (2005) Effect of herbicidal application of 2,4-dichlorophenoxyacetic acid in Arabidopsis. Funct Integr Genomics 5:4–17
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6:420–425
Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379:633–646
Roux F, Gasquez J, Reboud X (2004) The dominance of the herbicide resistance cost in several Arabidopsis thaliana mutant lines. Genetics 166:449–460
Sato T, Theologis A (1989) Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc Natl Acad Sci U S A 86:6621–6625
Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32:1011–1022
Schaller GE, Kieber JJ (2002) Ethylene. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, MD, pp 1–18
Schmidt R, Willmitzer L (1991) Arabidopsis regeneration and transformation (leaf and cotyledon explant system). Kluwer, Dordrecht, The Netherlands
Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K (2000) An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol 41:898–903
Taylor IB, Burbidge A, Thompson AJ (2000) Control of abscisic acid synthesis. J Exp Bot 51:1563–1574
Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M (1995) The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J 8:561–569
Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signalling networks. Plant Cell 14(Suppl):S131–S151
Wang W, Hall AE, O’Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A 100:352–357
Zarembinski TI, Theologis A (1994) Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol 26:1579–1597
Acknowledgements
We would like to thank Nufarm Limited, Melbourne, Victoria, Australia, for the financial support.
This work was supported by Nufarm and an RMIT University VRI grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Experiment station: Plant Biotechnology Centre, Primary Industries Research Victoria, Department of Primary Industries, La Trobe University, Bundoora, Victoria 3086, and the Victorian Microarray Technology Consortium (VMTC).
Rights and permissions
About this article
Cite this article
Raghavan, C., Ong, E.K., Dalling, M.J. et al. Regulation of genes associated with auxin, ethylene and ABA pathways by 2,4-dichlorophenoxyacetic acid in Arabidopsis . Funct Integr Genomics 6, 60–70 (2006). https://doi.org/10.1007/s10142-005-0012-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-005-0012-1