Abstract
The Pacific white shrimp Litopenaeus vannamei is a predominant aquaculture shrimp species in the world. Like other animals, the L. vannamei exhibited sexual dimorphism in growth trait. Mapping of the sex-determining locus will be very helpful to clarify the sex determination system and further benefit the shrimp aquaculture industry towards the production of mono-sex stocks. Based on the data used for high-density linkage map construction, linkage-mapping analysis was conducted. The sex determination region was mapped in linkage group (LG) 18. A large region from 0 to 21.205 cM in LG18 showed significant association with sex. However, none of the markers in this region showed complete association with sex in the other populations. So an association analysis was designed using the female parent, pool of female progenies, male parent, and pool of male progenies. Markers were de novo developed and those showing significant differences between female and male pools were identified. Among them, three sex-associated markers including one fully associated marker were identified. Integration of linkage and association analysis showed that the sex determination region was fine-mapped in a small region along LG18. The identified sex-associated marker can be used for the sex detection of this species at genetic level. The fine-mapped sex-determining region will contribute to the mapping of sex-determining gene and help to clarify sex determination system for L. vannamei.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Pacific white shrimp Litopenaeus vannamei (L. vannamei) is native to the Pacific coast of Mexico southward to Peru where the water temperatures normally higher than 20 °C throughout the year. As L. vannamei shows fast growth, relatively low protein requirements (20–25%), and ability to adapt well to variable environmental conditions, it is now becoming the predominant aquaculture shrimp farmed in China, Southeast Asia, South, and North America (Mathew Briggs et al. 2005). The total production was 3,668,681 t in 2014 which accounts for >70% of the worldwide farmed shrimp production (FAO). Like the other animals, the L. vannamei exhibited sexual dimorphism in growth trait (Gitterle et al. 2005). The female individuals are bigger than the males after sexual maturity (Pérez-Rostro CIaI, A.M. 2003). Clarification of the sex-determining system will not only enrich the knowledge in crustacean but also benefit the shrimp aquaculture industry towards the production of mono-sex stocks or sex-controlled stocks (Palaiokostas et al. 2013b).
Sex determination is an important area of study in developmental, evolutionary biology and ecology (Charlesworth and Mank 2010). The mechanisms of sex determination in animals are remarkably diverse and were controlled by genetic and/or environmental factor. The genetic mechanism of sex determination commonly shows either XX/XY (male heterogamety), or ZZ/ZW (female heterogamety) mechanism. In eutherian mammals, sex determination is defined by the male heterogametic XX/XY system (Charlesworth and Mank 2010); while in birds (Stevens 1997), lepidopteran insects (Sanchez 2008), and some teleost fish (Chen et al. 2014; Cnaani et al. 2008; Koyama et al. 2015), the sex determination system is principally female heterogametic ZW/ZZ system. In order to clarify the sex determination mechanism in a species, the first step is to map the sex-determining region. Previous reports showed that sex could be controlled by a major sex determination gene in one chromosome or multiple genes in different chromosomes (Gamble and Zarkower 2014). For penaeid shrimp (Decapoda), as non-environmental sex determination has been reported, the sex might be controlled mainly by genetic factor (Benzie 1998; Campos-Ramos et al. 2006; Legrand et al. 1987). The linkage-mapping analysis of Penaeus monodon, Penaeus japonicus, and L. vannamei detected major sex-determining QTLs in one LG (Alcivar-Warren et al. 2007; Du et al. 2010; Li et al. 2003; Pérez et al. 2004; Robinson et al. 2014; Staelens et al. 2008; Zhang et al. 2006), which give evidence to the existence of major sex-determining gene and sex chromosome. The sex-determining region of P. monodon was mapped in a narrow region on LG 30 based on the constructed high-density linkage map (Robinson et al. 2014); however, the marker showing the strongest association with sex was not completely linked to sex. Meanwhile, the sex-determining region in P. japonicus was only mapped in a large region of LG 28 and was still not fine-mapped (Li et al. 2003). Several studies have identified the QTLs of sex determination in L. vannamei, the mapped region was still almost 20 cM and none sex-specific marker was identified (Du et al. 2010). So, more detailed mapping studies are needed to identify the region and genes associated with sex determination.
Linkage mapping and association mapping are two major strategies to identify genetic loci and markers for interested traits. In previous research, mapping of sex-determining loci was conducted by AFLP, RAPD, or SSR markers. To map the sex-determining region or markers accurately, a large amount of markers were needed. With the development of next generation sequencing, markers genotyping such as SNPs and Indel became easier. Among the genotyping method based on next generation sequencing, one effective method was called SLAF-seq, which has proved to be effective in marker-genotyping and linkage map construction (Sun et al. 2013; Zhang et al. 2013). Recently, a high-density linkage map with 6146 marker mapped to 44 sex-averaged linkage groups has been constructed for L. vannamei in our group, the average marker distance was 0.7 cM (Yu et al. 2015).
The aim of the present study was to identify the sex-determining region and sex-specific marker for L. vannamei. Linkage mapping was performed based on the high-density linkage map constructed previously. Association analysis was conducted using the SLAF-seq data of two parents, 50 female progenies, and 50 male progenies. As a result, the sex-determining region was mapped to linkage group (LG) 18 and a sex marker was identified. The results support the female heterogametic (ZW) sex determination mechanism in L. vannamei and will contribute to the mapping of sex-determining genes in the further research.
Materials and Methods
Data Collection
The materials and the data used in this study were the same as previously reported (Yu et al. 2015). Briefly, a full-sib family including two parents and 205 offspring was collected. The sex of each individual was determined by the presence or absence of petasma on the first pleopods. Female individuals were scored as 1 and males were scored as 0. DNA from all these individuals was extracted for library construction. The sample DNA was firstly digested with MseI (New England Biolabs, (NEB)) and then digested with the additional restriction enzyme EcoRI and Nla III. PCR reactions were performed to amplify the digested DNA and barcodes were added to the different samples, then the DNA was pooled and used for library construction. The libraries were sequenced using paired-end sequencing method on Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, CA, USA).
Linkage-mapping Analysis
Based on the next generation sequencing data, a high-density linkage map was constructed. A total of 6146 markers spanning 4271.43 cM were mapped to 44 sex-averaged linkage groups, with an average marker distance of 0.7 cM (Yu et al. 2015). To identify regions of the genome that possess QTLs of sex, the composite interval mapping method applied in MapQTL5 software package was used (Van Ooijen 2004). The LOD score significance thresholds were determined by 1000 permutation tests for the sex trait. A QTL was determined to be significant if the LOD score was higher than the genome-wide significance threshold estimated by permutation.
Validation of Sex Markers Identified by Linkage Mapping
The association between the genotype of mapped markers and the phenotypic sex in the mapping family was re-analyzed using R/SNPassoc (Gonzalez et al. 2007). As the package can only handle the bi-allele markers, the three or four allele SLAF markers were discarded. The markers that are statistically significant after correcting for the number of tests performed (Bonferroni correction) were considered as sex-associated markers. Then, the significant markers detected by linkage mapping and the R/SNPassoc analysis were validated in different populations by Sanger sequencing. The marker sequences were blasted to the assembled genome scaffold of L. vannamei to obtain the sequence beside the markers. Primers were designed to successfully amplify and sequence the SNPs. The PCR profile included an initial 5 min of denaturation at 94 °C, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; and a final step of extension at 72 °C for 5 min. The PCR products were sequenced using ABI Prism 3730 (Applied Biosystems, USA) with the forward or reverse primer. The genotypes of candidate markers in female and male individuals were obtained by checking the Sanger sequence chromatogram at the SNP position. The populations used for verification made up of five commercial lines: SIS commercial line at the year of 2014 (Shrimp Improve System Co., USA), CP commercial line at the year of 2014 (Chia Tai Group, Thailand), Kona Bay commercial line at the year of 2014 (Kona Bay Marine Resources, USA), local selective breeding line selected from Hainan Guangtai Ocean Breeding Co., Ltd. (Hainan province, China), and shrimp individuals imported from Ecuador in 2014. Each line contained 12 female and 12 male individuals. The markers showed complete associations with sex among all the female and male individuals were considered as real sex-associated markers.
Association-mapping Analysis
An association-mapping method similar to bulked segregant analysis (BSA) was applied to further map the sex determination region and sex markers. Data from two parents and 50 female progenies and 50 male progenies with high-sequencing coverage were selected. These data was divided into four groups: female parent (M), pool of female progenies (aa), male parent (P), and pool of male progenies (ab).
De novo marker development was achieved by the following procedures. The paired sequences were combined by ten repeat of “X”. The trimmed SLAF pair-end reads were clustered based on the sequence similarity as detected by BLAT with the following parameters: “−tileSize = 10 and −stepSize = 5” (Kent 2002). Sequences with over 90% identity were grouped in one SLAF locus (Zhang et al. 2013). The SLAF loci were separated by the sequence differences into various SLAF tags. Those SLAF loci with more than four tags were filtered out because SLAF locus can contain no more than four genotypes in a mapping population of diploid species. The read depth of each SLAF tag in each individual or pool was calculated based on the mapping result.
For the polymorphic SLAF locus, read depth of each allele in the four datasets (M, aa, P, and ab) was obtained. The ratio of each allele read depth in female pool (M + aa) dividing male pool (P + ab) was calculated respectively. Then the markers were ranked by the ratio. The read depth in M and P dataset were also used to verify whether the marker was female or male-specific. For the non-polymorphic SLAF markers, similar analysis was conducted. Those sequences detected in M and aa with high read depth but not in P and ab were considered as female-specific locus, and vice versa for male-specific locus.
Validation of Sex Markers Identified by Association Mapping
The candidate sex markers identified by association analysis was first verified in mapping family using 30 female and 30 male progenies, and then were validated in five different populations. The procedure of validation was the same as that in linkage-mapping analysis.
Integration of Linkage and Association-mapping Results
In order to fine-map the sex-determining region in the LGs, besides the marker identified by linkage mapping, markers identified by association mapping were genotyped and mapped to the constructed linkage map. These markers were firstly blasted to the markers identified by linkage mapping to check whether there are same markers. Then the markers detected by association analysis were genotyped by ABI SNaPshot assay in the mapping family. The genotypes of these markers were joined with other data generated from high-throughput sequencing. In order to reduce computational complexity, only markers in the sex-associated LGs were selected. The integrated data were imported to JoinMap 4.0 (Kyasma, NL). A LOD threshold of 10.0 was used for assigning markers into different linkage groups. The Kosambi-mapping function based on regression mapping was used for map construction.
Results
Linkage Mapping of Sex-determining Region
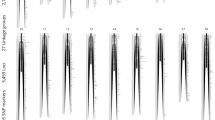
The genome-wide significance thresholds of LOD score detected by permutation test was 5.1. Results from the linkage-mapping analysis showed that a large region from 0 to 21.205 cM in LG18 showed significant association with sex determination (Table 1). The highest LOD score was 71.44, which was observed from 9.144 to 9.242 cM in LG18. Four adjacent markers (Marker13273, Marker16044, Marker23846, Marker12948) were located in this region. The LOD scores for the nearby markers including Marker728, Marker16149, Marker616, Marker6711, Marker1962, Marker13107, and Marker23573 were also high to 71.43, which were very similar to the peak scores (Fig. 1).
Validation of Candidate Sex Markers Identified by Linkage Mapping
After Bonferroni correction, ten markers in LG18 showed significant associations with sex in the mapping family (Supplementary Table S1). The most significant markers were Marker728 and Marker16149, which were located at 8.605 cM of LG18 (Fig. 2). The other eight markers were located in a region ranged from 2.907 to 15.711 cM. Combining the linkage analysis and association analysis using R/SNPassoc, the most significantly associated region with sex in LG18 was located at 8.605 cM.
These significant markers detected by the two methods were validated in different populations. However, none of them showed complete association with sex determination.
Association Mapping of Sex-associated Markers
As no marker identified by linkage mapping was fully linked with sex in the validated populations, we applied an association analysis approach to further map the sex determination region and sex markers. The association analysis was similar to bulk segregant analysis (BSA) analysis. A total of 8,840,934 paired-end reads from the female parent, 11,626,188 paired-end reads from the male parent, 129 million reads from female offspring pool, and 126 million from male offspring pool were used for further analysis. Based on these data, SLAF markers were developed separately for the four datasets. As a result, 89,699 markers were generated for the female parent and 94,256 markers for the male parent. As the female and male progeny pool contained much more reads, more markers were obtained, including 149,529 markers from the pool of female progenies and 149,659 markers from the pool of male progenies.
The read depth of markers in each dataset was calculated based on the mapping result. For the polymorphic markers, three markers showing significant differences between female and male were identified (Table 2). All the three markers contained two alleles. One allele could be observed both in female and male pool with similar read depth. The other allele showed significantly higher read depth in female than male. For the non-polymorphic markers, a total of 13 candidate sex markers were observed. Among them, 12 markers showed significantly higher read depth in female pool than that in male pool, the other one marker was only observed in male pool (Table 3). All the detected sex-associated markers were displayed in Tables S2 and S3.
Validation of Candidate Sex Markers Identified by Association Mapping
Among the markers identified by association analysis, all could find the hit to the assembled genome scaffold of L. vannamei except for Marker41731, Marker10360, Marker36508, and Marker 99554. Interestingly, the non-polymorphic markers associated with sex, which was detected in one gender in association analysis, could be amplified in both genders in the individuals of the mapping family. Through Sanger sequencing of these amplified fragments, we identified SNPs in the enzyme sites, which showed differences between female and male.
After verification of these candidate markers in the mapping family, we found that three markers including Marker24863, Marker24577, and Marker19299 showed significant difference between female and male individuals in the mapping family (Supplementary Figs. S1, S2, and S3). The primer sequence designed for amplifying these three marker sequences was displayed in Table S4. For Marker19299, the sequence difference between female and male was located at the position of 20 bp in Fig. S3. However, it was not completely different between female and male. So, we further checked the sequencing chromas. Interestingly, the nucleotide at the position of 80 bp of Marker19299 showed a pattern that all females were C/G heterozygotes and all the males were C homozygotes. Another mutation of Marker19299 was observed at 284 bp, there was a 9-bp length insert in female compared to male sequence. The existence of this insert resulted in the sequencing error for the behind sequence. The sequencing chromas of Marker24863 and Marker24577 were checked and they also showed female heterogamety and male homogamety.
The validation of all the identified markers in different populations showed that Marker19299 was completely associated with sex trait. At the 80 bp of the Marker19299 sequence in Fig. S3, all the female individuals were C/G heterozygotes and all the male individuals were C homozygotes. Although the other markers showed the trend of female heterogamety and male homogamety, the pattern was not applicable to all the females and males. This result indicated that Marker19299 was a real sex marker.
Integration of Linkage and Association-mapping Results
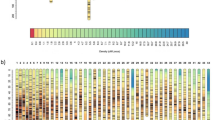
After blasting markers identified from association analysis to those in linkage analysis, it showed that Marker41731 in association analysis was the same as Marker65257 in linkage analysis. In the linkage map constructed previously, Marker65257 was located at 1.106 cM of LG 18, which was a QTL for sex with high LOD score. The reconstruction of sex-associated LGs after adding Marker24863, Marker24577, and Marker19299 reveals that all markers except for Marker24577 were mapped to LG 18 (Fig. 3). High degree of collinearity between the reconstructed map and the previous map was observed (Fig. 4). In the LG 18 of the reconstructed map, Marker19299 was mapped to 8.955 cM, which was 1.751 cM away from Marker16149 and Marker728, and it was 1.4 cM away from Marker1297 (Fig. 3). As Marker16149 and Marker728 were the most significant markers in the mapping family, the sex-determining region should be located between Marker19299 and Marker16149 (Marker728). Since the genome assembling of L. vannamei was not finished yet, the sequence between these markers was not fully assembled and annotated.
Discussion
In the present study, linkage mapping and association mapping analyses were applied to identify QTLs for sex determination and the corresponding sex-associated markers. Linkage mapping analysis identified a major QTL in LG18. The association analysis identified three sex-associated markers, among which one marker was fully sex-associated. The integration of linkage and association mapping precisely mapped the sex-determining region in a narrow region along LG 18. This study was the first report to map QTL of sex determination in a narrow region for L. vannamei. The finding in this study will be useful for mapping of the sex-determining genes and revealing the sex chromosome characteristics in the future study.
Previously, screening of sex-specific markers was performed using random amplified polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP) approach (Pan et al. 2015; Vale et al. 2014; Ventura et al. 2011). Recently, next generation sequencing has allowed to genotype thousands of markers simultaneously in a short time. Genotyping-based next generation sequencing has been proved effective in genetic variation discovery (DePristo et al. 2011; Kerstens et al. 2009; Yu et al. 2014), linkage map construction (Li et al. 2014; Shao et al. 2015; Wu et al. 2014), population genetic research (Fumagalli et al. 2013; Larson et al. 2014), and association mappings (Houston et al. 2012; Kim et al. 2010). So, many studies focused on the mapping of sex determination QTLs using the next generation sequencing platform. Researchers identified sex-linked SNP markers using RAD sequencing and supported ZW/ZZ sex determination mechanism in Pistacia vera L. (Kafkas et al. 2015). In another study, seven male and ten female Anolis carolinensis were sequenced using RAD-seq technology, one male-specific molecular marker was identified (Gamble and Zarkower 2014). In aquaculture, most of the similar study were reported in fish, including Nile tilapia (Palaiokostas et al. 2013b), Atlantic halibut (Palaiokostas et al. 2013a), tiger pufferfish (Kamiya et al. 2012), and zebrafish (Anderson et al. 2012). These studies identified QTLs for sex and male-specific markers, which supported the XX/XY sex determination system in these fish species. Among crustaceans, the sex marker of salmon louse, which was one of the major ectoparasitisms occurred on marine salmonids, has been identified through RAD sequencing. But for penaeid, this study is the first report to identify sex-specific markers using next generation sequencing. Our result illustrated the efficiency of this method for identifying QTLs and the sex-associated marker. The data of the female heterogamety shown by sex-associated markers give more evidence to support the ZW/ZZ sex determination mechanism in L. vannamei.
To map QTLs for sex and identify sex-associated marker, most of the study used family material and some use unrelated individuals collected from different populations. Usually, the family material was firstly used to map the sex loci in the chromosome, and then the mapped loci were validated in the other individuals, like what was performed in this study and the research of yellowtail, pistacia, and Atlantic halibut (Kafkas et al. 2015; Koyama et al. 2015; Palaiokostas et al. 2013a). Recently, as the development of marker-genotyping, researchers proposed an approach to sequence the unrelated individuals from natural populations to screen the sex-associated markers (Gamble and Zarkower 2014). One advantage of this method is that there’s no need to perform experimental crosses and construct families, and the materials were easy to be collected. In our opinion, both the methods were efficient in identifying sex-specific markers. For species without genome reference, it would be better to map the sex-determining gene using family material. For species with its reference genome available, mapping of the sex-determining gene can be achieved directly by genome-wide association study (GWAS) using different populations.
Linkage analysis, as well as association analysis, is a high-effective way for identifying the genome position which controls the phenotype. In the present study, both methods were applied to identify the sex-determining region and sex markers in the L. vannamei. Linkage analysis found the sex-associated chromosomes and identified the loci significantly linked with sex. At the beginning, a candidate sex chromosome was detected by linkage analysis. Unfortunately, none of the markers identified by linkage mapping could be used as sex marker in the other populations. So, an association mapping approach similar to BSA analysis was performed. More sex-associated markers were identified, including one marker fully associated with sex. Most of the markers detected in association analysis were not identified in the linkage analysis. One reason was that the marker development procedure in linkage analysis was only conducted in parents (Yu et al. 2015) while the marker identified in the association analysis was performed in both parents and pool of offspring, and the high read depth in the pool of offspring could lead to more markers. Another reason was that the markers mapped in the linkage map should be successfully genotyped in at least 75% of the offspring, the markers genotyped in less than 75% of the offspring will not be appeared in the linkage map, and these markers will be discarded from the linkage analysis. Our study illustrated that the integration of linkage and association analyses could make good use of the data generated from the next generation sequencing and could assist to fine map the sex-determining region.
In the previous cytological study, no heteromorphic sex chromosomes were observed in L. vannamei (Campos-Ramos 1997). It indicated that the sex chromosome pair was very similar in female and male. The large region of specific sequence in one sex may not be existed in shrimp. The fact that there are no heteromorphic sex chromosomes indicated that the sex chromosome of shrimp was young. The sex chromosome pair has not diverged yet (Charlesworth and Mank 2010). So the non-recombining region in this species may be small. Besides, the recombination rate was reported to be high in L. vannamei (Zhang et al. 2006); the detected sex-linked markers may cross over frequently during meiosis. So, the linkage disequilibrium of the identified sex-linked markers with the sex-determining genes may disappear in the other population. This can partly explained why some of the screened sex-linked markers in family were not applicable in the other populations. Nevertheless, the Marker19299 was detected fully associated with sex both in family and populations. This sex marker was mapped to the most significant region-associated with sex in the LG18, which as a result fine-mapped the sex-determining loci in a region of 1.751 cM. To our knowledge, this is the most precise sex-determining region for L. vannamei. With the help of genome sequencing, the fine-mapping of sex-determining genes and clarifying sex determination mechanism of L. vannamei will be possible in the near future.
Conclusions
Overall, this study successfully mapped sex-determining region and identified a marker completely associated with sex by linkage and association analyses. The sex-determining region was fine-mapped in the LG 18. The sex marker showed female heterogamety, which supported the hypothesis of ZW/ZZ sex determination system in L. vannamei. The identified sex-associated marker could be of high value towards the determination of genetic sex and the production of mono-sex stocks or sex-controlled stocks. These findings will contribute to an improved understanding of sex determination and the further fine-mapping of sex-determining genes for L. vannamei.
References
Alcivar-Warren A, Meehan-Meola D, Park SW, Xu Z, Delaney M, Zuniga G (2007) ShrimpMap: a low-density, microsatellite-based linkage map of the pacific whiteleg shrimp, Litopenaeus vannamei: identification of sex-linked markers in linkage group 4. J Shellfish Res 26:1259–1277
Anderson JL, Rodriguez Mari A, Braasch I, Amores A, Hohenlohe P, Batzel P, Postlethwait JH (2012) Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One 7:e40701
Benzie JAH (1998) Penaeid genetics and biotechnology. Aquaculture 164:23–47
Briggs M, Funge-Smith S, Subasinghe RP, Phillips M (2005) Introductions and movement of two penaeid shrimp species in Asia and the Pacific. Food and Agriculture Organization of the United Nations. FAO Fisheries Technical Paper 476
Campos-Ramos R, Garza-Torres R, Guerrero-Tortolero DA, Maeda-Martinez AM, Obregon-Barboza H (2006) Environmental sex determination, external sex differentiation and structure of the androgenic gland in the Pacific white shrimp Litopenaeus vannamei (Boone). Aquac Res 37:1583–1593
Campos-Ramos R (1997) Chromosome studies on the marine shrimps Penaeus vannamei and P-californiensis (Decapoda). J Crustac Biol 17:666–673
Charlesworth D, Mank JE (2010) The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186:9–31
Chen SL et al (2014) Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46:253–260
Cnaani A et al (2008) Genetics of sex determination in tilapiine species. Sex Dev 2:43–54
DePristo MA et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498
Du ZQ, Ciobanu DC, Onteru SK, Gorbach D, Mileham AJ, Jaramillo G, Rothschild MF (2010) A gene-based SNP linkage map for pacific white shrimp, Litopenaeus vannamei. Anim Genet 41:286–294
Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sanchez E, Albrechtsen A, Nielsen R (2013) Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195:979–992
Gamble T, Zarkower D (2014) Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol Ecol Resour 14:902–913
Gitterle T et al (2005) Genetic (co)variation in harvest body weight and survival in Penaeus (Litopenaeus) vannamei under standard commercial conditions. Aquaculture 243:83–92
Gonzalez JR, Armengol L, Sole X, Guino E, Mercader JM, Estivill X, Moreno V (2007) SNPassoc: an R package to perform whole genome association studies. Bioinformatics 23:644–645
Houston RD et al (2012) Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics 13:244
Kafkas S, Khodaeiaminjan M, Guney M, Kafkas E (2015) Identification of sex-linked SNP markers using RAD sequencing suggests ZW/ZZ sex determination in Pistacia vera L. BMC Genomics 16:98
Kamiya T et al (2012) A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet 8:e1002798
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kerstens HHD, Crooijmans RPMA, Veenendaal A, Dibbits BW, Chin-A-Woeng TFC, den Dunnen JT, Groenen MAM (2009) Large scale single nucleotide polymorphism discovery in unsequenced genomes using second generation high throughput sequencing technology: applied to turkey. BMC Genomics 10:479
Kim SY et al (2010) Design of association studies with pooled or un-pooled next-generation sequencing data. Genet Epidemiol 34:479–491
Koyama T et al (2015) Identification of sex-linked SNPs and sex-determining regions in the yellowtail genome. Mar Biotechnol (NY) 17:502–510
Larson WA, Seeb LW, Everett MV, Waples RK, Templin WD, Seeb JE (2014) Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol Appl 7:355–369
Legrand JJ, Legrandhamelin E, Juchault P (1987) Sex determination in Crustacea. Biol Rev 62:439–470
Li B et al (2014) Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max. BMC Genomics 15:1086
Li YT et al (2003) Genetic mapping of the kuruma prawn Penaeus japonicus using AFLP markers. Aquaculture 219:143–156
Pérez-Rostro CI, Ibarra AM (2003) Heritabilities and genetic correlations of size traits at harvest in sexually dimorphic Pacific white shrimp (Litopenaeus vannamei) grown in two environments. Aquac Res 34:1079–1085
Pérez F, Erazo C, Zhinaula M, Volckaert F, Calderón J (2004) A sex-specific linkage map of the white shrimp Penaeus (Litopenaeus) vannamei based on AFLP markers. Aquaculture 242:105–118
Palaiokostas C et al (2013a) Mapping the sex determination locus in the Atlantic halibut (Hippoglossus hippoglossus) using RAD sequencing. BMC Genomics 14:566
Palaiokostas C, Bekaert M, Khan MGQ, Taggart JB, Gharbi K, McAndrew BJ, Penman DJ (2013b) Mapping and validation of the major sex-determining region in Nile tilapia (Oreochromis niloticus L.) using RAD sequencing. PLoS One 8(7):e68389
Pan ZJ, Li XY, Zhou FJ, Qiang XG, Gui JF (2015) Identification of sex-specific markers reveals male heterogametic sex determination in Pseudobagrus ussuriensis. Mar Biotechnol (NY) 17:441–451
Robinson NA et al (2014) QTL for white spot syndrome virus resistance and the sex-determining locus in the Indian black tiger shrimp (Penaeus monodon). BMC Genomics 15:731
Sanchez L (2008) Sex-determining mechanisms in insects. Int J Dev Biol 52:837–856
Shao C et al (2015) Genome-wide SNP identification for the construction of a high-resolution genetic map of Japanese flounder (Paralichthys olivaceus): applications to QTL mapping of Vibrio anguillarum disease resistance and comparative genomic analysis. DNA Res 22:161–170
Staelens J, Rombaut D, Vercauteren I, Argue B, Benzie J, Vuylsteke M (2008) High-density linkage maps and sex-linked markers for the black tiger shrimp (Penaeus monodon). Genetics 179:917–925
Stevens L (1997) Sex chromosomes and sex determining mechanisms in birds. Sci Prog 80(Pt 3):197–216
Sun XW et al (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8(3):e58700
Vale L, Dieguez R, Sanchez L, Martinez P, Vinas A (2014) A sex-associated sequence identified by RAPD screening in gynogenetic individuals of turbot (Scophthalmus maximus). Mol Biol Rep 41:1501–1509
Van Ooijen JW (2004) MapQTL 5. Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, Netherlands
Ventura T, Aflalo ED, Weil S, Kashkush K, Sagi A (2011) Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity (Edinb) 107:456–461
Wu J et al (2014) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot 65:5771–5781
Yu Y, Wei JK, Zhang XJ, Liu JW, Liu CZ, Li FH, Xiang JH (2014) SNP discovery in the transcriptome of white Pacific shrimp Litopenaeus vannamei by next generation sequencing. PLoS One 9(1):e87218
Yu Y et al (2015) Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific white shrimp Litopenaeus vannamei. Sci Rep 5:15612
Zhang L, Yang C, Zhang Y, Li L, Zhang X, Zhang Q, Xiang J (2006) A genetic linkage map of Pacific white shrimp (Litopenaeus vannamei): sex-linked microsatellite markers and high recombination rates. Genetica 131:37–49
Zhang Y et al (2013) Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol 13:141
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work is supported by National Natural Science Foundation of China (Grant Nos. 31502161 and 31302171, the Joint NSFC-ISF Research Program (31461143007), China Agriculture Research System-47 (CARS-47), and the Scientific and Technological Innovation Project Financially Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2015ASKJ02).
Conflict of Interest
The authors declare that they have no competing interests.
Electronic supplementary material
Supplementary Fig. S1
Demonstration of sequence difference of Marker24863 between female and male individuals in the mapping family. (GIF 127 kb)
Supplementary Fig. S2
Demonstration of sequence difference of Marker24577 between female and male individuals in the mapping family. (GIF 144 kb)
Supplementary Fig. S3
Demonstration of sequence difference of Marker19299 between female and male individuals in the mapping family. (GIF 176 kb)
Supplementary Table S1
Association analysis of candidate sex markers with phenotypic sex trait using R/SNPassoc in the mapping family. (XLSX 10 kb)
Supplementary Table S2
Marker name and sequences of candidate sex-associated polymorphic SLAF markers. (XLSX 9 kb)
Supplementary Table S3
Marker name and sequences of candidate sex-associated non-polymorphic SLAF markers. (XLSX 10 kb)
Supplementary Table S4
Primer sequences for amplifying the sex-linked markers identified by association analysis. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Yu, Y., Zhang, X., Yuan, J. et al. Identification of Sex-determining Loci in Pacific White Shrimp Litopeneaus vannamei Using Linkage and Association Analysis. Mar Biotechnol 19, 277–286 (2017). https://doi.org/10.1007/s10126-017-9749-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-017-9749-5