Abstract
A new lectin from Aplysia dactylomela eggs (ADEL) was isolated by affinity chromatography on HCl-activated Sepharose™ media. Hemagglutination caused by ADEL was inhibited by several galactosides, mainly galacturonic acid (Ka = 6.05 × 106 M−1). The primary structure of ADEL consists of 217 residues, including 11 half-cystines involved in five intrachain and one interchain disulfide bond, resulting in a molecular mass of 57,228 ± 2 Da, as determined by matrix-assisted laser desorption/ionization time of flight mass spectrometry. ADEL showed high similarity with lectins isolated from Aplysia eggs, but not with other known lectins, indicating that these lectins could be grouped into a new family of animal lectins. Three glycosylation sites were found in its polypeptide backbone. Data from peptide-N-glycosidase F digestion and MS suggest that all oligosaccharides attached to ADEL are high in mannose. The secondary structure of ADEL is predominantly β-sheet, and its tertiary structure is sensitive to the presence of ligands, as observed by CD. A 3D structure model of ADEL was created and shows two domains connected by a short loop. Domain A is composed of a flat three-stranded and a curved five-stranded β-sheet, while domain B presents a flat three-stranded and a curved four-stranded β-sheet. Molecular docking revealed favorable binding energies for interactions between lectin and galacturonic acid, lactose, galactosamine, and galactose. Moreover, ADEL was able to agglutinate and inhibit biofilm formation of Staphylococcus aureus, suggesting that this lectin may be a potential alternative to conventional use of antimicrobial agents in the treatment of infections caused by Staphylococcal biofilms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lectins are sugar-binding proteins that possess the ability to decipher the glycocode. They have been found in all living organisms. In mollusks, lectins have been detected and purified from cephalopods, gastropods, and bivalves (Alpuche et al. 2010; Bulgakov et al. 2004; Fujii et al. 2011; Song et al. 2011). Among the prospected lectins representatives of some structural families are found, such as C-, F-, and R-types; galectins; and RBLs (Chen et al. 2011; Fujii et al. 2012; Naganuma et al. 2006; Pales-Espinosa et al. 2010; Song et al. 2011).
The presence of lectins in mollusks has been associated with hemolymph, mucus, hemocytes, and reproductive tissues (Chen et al. 2011; Gilboa-Garber et al. 1985; Ito et al. 2011; Zhang et al. 2014), and evidences suggest that they can act as pattern recognition receptors (PRRs), recognizing several pathogen-associated molecular patterns (PAMPs). In fact, lectin-coding genes have been found be upregulated after infection by pathogenic microorganisms (Chen et al. 2011; Mu et al. 2014). Moreover, some studies have explicitly demonstrated the antibacterial potential of lectins isolated from mollusks (Zheng et al. 2008; Takahashi et al. 2008; He et al. 2011).
Gastropoda is the most broadly distributed class of mollusk, and several lectins from gastropods have been isolated (Fujii et al. 2011; Gilboa-Garber et al. 1985; Kawsar et al. 2009). Aplysia dactylomela Rang, 1828 is a circumtropical aplysid gastropod and occurs worldwide in tropical to warm temperate waters (Matthews-Cascon et al. 2011). In the West Atlantic, this species is found in Florida, Mexico, the Caribbean, and Brazil. It is the biggest opisthobranch found in Northeastern Brazil and one of the main herbivores inhabiting consolidated intertidal bedrock. The egg mass of an A. dactylomela is a resistant gelatinous string that can reach 8 m in length. Egg mass color is variable, reaching tones of yellow, orange, and pink (Matthews-Cascon et al. 2011).
Currently, only a fetuin-binding lectin named dactylomelin-P has been isolated and characterized in some detail from the purple gland of A. dactylomela (Melo et al. 2000).
In the present work, we have isolated, characterized, and determined the primary structure and estimated the secondary structure of a new lectin from the eggs of A. dactylomela. Additionally, we created a 3D model and evaluated its antibacterial and antibiofilm potentials.
Material and Methods
Egg Collection and Extraction
Aplysia dactylomela eggs were collected during low tide in the intertidal zone at Pacheco Beach (3° 41′ 13.0″ S; 38° 38′ 21.7″ W), Caucaia, Brazil. Collections were authorized and certified by responsible environmental authorities (SISBIO ID: 33913-8).
Eggs were removed from stones and algae, washed with seawater, and transported to the lab. Immediately thereafter, eggs were crushed using mortar and pestle. Proteins from smashed eggs were extracted with ten volumes of 20 mM Tris-HCl, pH 7.6, containing 150 mM NaCl and phenylmethylsulfonyl fluoride (PMSF) 0.1 mM (Tris-buffered saline (TBS)). The mixture was then centrifuged at 8000×g for 20 min at 4 °C. The crude extract was stored at −20 °C until use. Protein content was determined by Bradford reagent, and BSA was used as protein standard (Bradford 1976).
Lectin Purification
The crude extract was applied to an HCl-activated Sepharose™ 4B column (1.0 × 8.0 cm), previously equilibrated with TBS. Unbound proteins were washed with the same buffer, and adsorbed proteins were recovered with TBS containing 200 mM α-lactose. Chromatography eluates were monitored by absorbance at 280 nm using a UV/Visible spectrophotometer. Retained fractions containing the purified lectin were exhaustively dialyzed against distilled water, freeze-dried, and stored at −20 °C until required.
Hemagglutinating Activity and Inhibition Assays
Hemagglutinating activity (HA) and inhibition assays were performed following pre-established methods (Sampaio et al. 1998), using human (A, B, and O) and rabbit erythrocytes, both treated with proteases and untreated.
The following sugars and glycoproteins were used in the inhibition assay: D-xylose, D-ribose, L-fucose, L-arabinose, L-rhamnose, D-galactose (Gal), D-mannose, D-glucose (Glc), D-glucosamine, D-galactosamine, N-acetyl-D-glucosamine (GlcNAc), N-acetyl-D-galactosamine (GalNAc), D-galacturonic acid, D-fructose, D-sucrose, D-melibiose, α-D-lactose, β-D-lactose, D-lactulose, D-maltose, D-raffinose, methyl-α-D-galactopiranoside, methyl-β-D-galactopiranoside, methyl-β-D-thiogalactose, phenyl-β-D-galactopiranoside, 4-nitrophenyl-α-D-galactopiranoside, 4-nitrophenyl-β-D-galactopiranoside, 2-nitrophenyl-β-D-galactopiranoside, O-nitrophenyl-β-D-galactopiranoside, and type 2 porcine stomach mucin (PSM).
Biochemical Characterization
The effects of pH, temperature, divalent cations, and EDTA on HA were determined as described by Sampaio et al. (1998). Molecular mass of the lectin was estimated by SDS-PAGE, as described by Laemmli (1970). LMW-SDS marker kit (GE Healthcare, IL, USA) was used as standard.
Native molecular mass was estimated by size exclusion chromatography (SEC) on BioSuite 250 HR SEC column (0.78 × 30 cm, 5-μm particle size, Waters Corp., MA, USA), coupled to an Acquity UPLC system (Waters Corp., MA, USA), which had been equilibrated with Tris-HCl 20 mM, pH 7.2, containing NaCl 500 mM. The column was previously calibrated with a mixture of standard proteins: conalbumin (75 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), ribonuclease A (14 kDa), and aprotinin (6.5 kDa).
Molecular Mass Determination
Molecular mass was determined on an AutoFlex III matrix-assisted laser desorption/ionization time of flight (MALDI-TOF)/TOF mass spectrometer (Bruker Daltonics, Germany). The samples were dissolved in deionized water at variable concentration, mixed (1:3 v/v) in an α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (5 mg/mL of CHCA on acetonitrile, water, and trifluoracetic acid, 50, 40, and 10% v/v), spotted onto a sample plate, and then dried at room temperature. The spectra were acquired in linear positive mode and were processed with Flex Analysis 3.4 software (Bruker Daltonics, Germany).
N-Terminal Sequencing
An aliquot of 15 μg of purified lectin was solubilized in 15 μL of acetonitrile 10% and centrifuged at 8000×g for 5 min at room temperature. The solution was applied into a glass fiber disk and submitted to N-terminal sequencing.
Automated Edman degradations were performed in the Shimadzu model PPSQ-31A protein sequencer (Shimadzu Corp., Japan). PTH-amino acids from the N-terminus sequence were separated on a 2.0 × 250 mm Wakosil ODS column (Wako Pure Chemical Corp., Osaka, Japan) connected to a model LC-20AT pump. The absorbance was detected at 269 nm with a UV–Vis SPD-20A detector.
Primary Structure Determination by Tandem Mass Spectrometry
Purified lectin was submitted to SDS-PAGE as described above. After staining, electrophoretic bands were excised using a plastic pipette tip. Spots were reduced with dithiothreitol, and the lectin was carboxamidomethylated (CAM-lectin) with iodoacetamide, following pre-established methods (Shevchenko et al. 2006).
Treated spots were digested with trypsin, chymotrypsin, and pepsin. Digestions with trypsin and chymotrypsin were performed in 50 mM ammonium bicarbonate at 1:50 w/w (enzyme/substrate). Digestion with pepsin was performed in 100 mM HCl at 1:50 w/w (enzyme/substrate). All digestions were maintained for 18 h and then stopped with 2 μL of 2% formic acid. Peptides were extracted from gel according to Shevchenko et al. (2006).
Peptide solution was loaded onto a C-18 (0.075 × 100 mm) nanocolumn coupled to a nanoAcquity system (Waters Corp., MA, USA). The eluates were directly infused into a nanoelectrospray source. Chromatography and mass spectrometry analysis were performed according to established methods (Carneiro et al. 2013).
Deglycosylation of tryptic peptides was carried out by Glycoprofile™ II, Enzymatic In-Solution N-Deglycosylation Kit (Sigma-Aldrich, St. Louis, MO, USA). First, the lectin (1 mg/mL) was solubilized in RapiGest™ SF surfactant 0.1% (Waters Corp., MA, USA), followed by digestion of CAM-lectin with trypsin 1:100 w/w (enzyme/substrate) for 18 h at 37 °C. RapiGest™ SF surfactant was pelleted by addition of TFA at final concentration of 0.2%.
Tryptic peptides were digested with peptide-N-glycosidase F (PNGase F) (1 U) (Sigma-Aldrich, St. Louis, MO, USA) for 18 h at 37 °C. Deglycosylated peptides were loaded onto a C-18 nanocolumn, and the eluates were directly infused into a nanoelectrospray source. The mass spectrometer was operated in positive mode with a source temperature of 373 K and a capillary voltage at 3.0 kV. MS/MS was performed according to the data-dependent acquisition (DDA) method. The lock mass used in acquisition was m/z 785.84 ion of the [Glu1] fibrinopeptide B. The selected precursor ions were fragmented by collision-induced dissociation (CID) using argon as the collision gas. All CID spectra were manually interpreted.
Circular Dichroism
Circular dichroism spectroscopic measurement was performed on a Jasco J-815 spectropolarimeter (Jasco International Co., Tokyo, Japan) connected to a peltier with controlled temperature. The lectin (0.2 mg/mL in 20 mM phosphate buffer, pH 7.0, containing 150 mM NaCl) was placed in a rectangular quartz cuvette with 0.5-mm path length. Spectra were acquired at a scan speed of 50 nm/min with a bandwidth of 1 nm. The acquisitions were performed at 190–250 nm (far-UV) and different temperatures (20–80 °C) with eight accumulations per temperature. The analysis of structural data was performed by DICHROWEB web server (Whitmore and Wallace 2008).
For measurement at 250–330 nm (near-UV), lectin at 0.5 mg/mL was previously incubated with D-galactose, α-D-lactose, D-galactosamine, and galacturonic acid at 10 mM each and placed in a rectangular cuvette with 5-mm path length. Spectra were acquired as described above.
The thermodynamic parameters of lectin folding and unfolding in the presence and absence of ligands were calculated by monitoring the changes in ellipticity at 293 nm as a function of temperature (Greenfield 2007). The lectin (8 μM) was submitted to temperatures ranging from 70 to 95 °C with a ramp rate of 4 °C/min, and sampling was conducted at 1 °C each in the presence and absence of D-galactose, D-galacturonic acid, D-galactosamine, and α-lactose at 80 μM each.
As proposed by Greenfield (2006, 2007), ligand-induced changes in the molar ellipticities of the lectin at 368 K were employed to find the affinity constant (Ka) for binding of carbohydrates to the lectin.
Structural Modeling
Structural Prediction
The structural prediction of the lectin was performed through interplay among the MODELER (Webb and Sali 2014) suite, I-TASSER (Yang and Zhang 2015), and QUARK (Xu and Zhang 2013) servers.
Initially, the complete protein sequence (217 amino acids) was submitted to I-TASSER and QUARK servers for overall modeling. In parallel, three sequence fragments from the lectin (amino acid residues 1–70, 71–141, and 142–217) were also submitted to these servers in order to obtain the individual folding of each protein domain. The best models were selected based on their C- and TM-scores2. Afterwards, the selected models were used as input to the MODELER suite, v.9.16. For structural modeling of the lectin, MODELER default parameters were used, and sequence alignment corrections were manually edited.
Initially, 30 theoretical models were generated, which were ranked based on their discrete optimized protein energy (DOPE) scores (Shen and Sali 2006). The models were then selected and analyzed for their stereochemical properties with the Protein Structure Validation Suite (PSVS) server, which integrates consolidated validation platforms, such as Procheck (Laskowski et al. 1993) and Molprobity (Davis et al. 2004). Finally, the best model was submitted to ModRefiner (Xu and Zhang 2013) (http://zhanglab.ccmb.med.umich.edu/ModRefiner).
Binding Site Prediction
A meta-server approach (http://zhanglab.ccmb.med.umich.edu) using the COACH (Yang et al. 2013) server was used to identify the most reliable carbohydrate-binding site.
Molecular Docking Calculations
The structural basis for carbohydrate recognition was explored through molecular docking calculations. For this purpose, four carbohydrates previously analyzed on hemagglutination inhibition assays were selected, including D-galactose (CID: 6036), D-galactosamine (CID: 24154), D-galacturonic acid (CID: 439215), and α-lactose (CID: 84571). All carbohydrate structures were obtained from the PubChem Substance and Compound database (Wang et al. 2009).
Calculations were performed with AutoDock Vina, version 1.1.2 (Trott and Olson 2009). The Autodock graphical interface AutoDockTools, version 1.5.6, was used to keep polar hydrogens and add partial charges to the proteins and ligands using Kollman United charges (Morris and Huey 2009). The protein and carbohydrate ligands were treated as rigid and flexible molecules, respectively.
Initially, a blind docking strategy was applied using a search space defined by a 40 Å × 60 Å × 40 Å cube covering the whole protein surface. Then, a refined search was performed using a search space defined by a 20 Å × 20 Å × 20 Å cube centered on the spot previously predicted by the COACH server as the most suitable carbohydrate-binding site. For all calculations, exhaustiveness was set to 15, and all other parameters were used as default. For each docking, the ten top-ranked generations based on the predicted binding affinity (in kilocalories per mole) were analyzed. The solutions were first chosen based on the presence of important interactions commonly visualized for carbohydrate recognition by lectins, such as the stacking of pyranosidic rings against aromatic or aliphatic amino acid residues and the proper disposal of hydrogen bonds around hydroxyl groups (Wittmann and Pieters 2013). Since all selected ligands presented similar chemical structures, a common binding pattern was sought. The most suitable results were further ranked based on the theoretical binding energy, which was given as a negative score in kcal/mol.
Antibacterial Activity
Microorganisms and Culture Conditions
The assays to evaluate antimicrobial activity were performed on Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 11303. Bacteria were grown in trypticase soy agar medium (TSA; Himedia, India) and incubated at 37 °C for 24 h. After growth on the solid medium, some isolated colonies were removed, inoculated into 10 mL trypticase soy broth (TSB; Himedia, India), and incubated for 24 h at 37 °C under constant agitation. Prior to use, the cell density of each bacterial suspension was adjusted to a final concentration of 2 × 106 cells/mL.
Bacterial Aggregation Assay
After bacterial growth, bacterial suspensions were harvested by centrifugation at 2000×g for 10 min, washed twice with TBS/Ca2+, suspended in TBS/Ca2+ containing formaldehyde 4% and then kept for 16 h at 4 °C. Afterwards, the cells were washed twice with TBS/Ca2+, and suspended in the same buffer. Bacterial count was estimated by the serial dilution method, and optical density at 620 nm (OD620) was maintained around 1.0. Bacterial agglutination was performed as described by Melo et al. (2014).
Effects of the Lectin on Planktonic Cells
The effects of lectin on planktonic cells were assessed by the broth microdilution method. Briefly, the lectin was diluted in 150 mM NaCl at concentrations ranging from 7.8 to 250 μg/mL. The assay was performed in 96-well polystyrene plates by addition of 100 μL of each bacterium at 2 × 106 cells/mL to each well. The microplates were then incubated at 37 °C for 24 h in constant agitation, and the optical density was measured at 620 nm using a microplate reader (SpectraMax® I3, Molecular Devices LLC, CA, USA).
Biofilm Formation Assay
The effects on biofilm formation were evaluated as proposed by Vasconcelos et al. (2014). Briefly, sterile 96-well polystyrene plates were prepared following the same steps as previously established for planktonic cell growth. However, two plates were used for this purpose: one for biomass quantification by crystal violet and the other for enumeration of biofilm viable cells. In order to verify the role of the carbohydrate recognition domain (CRD) on antibiofilm activity exerted by the lectin, it was tested after previous incubation with 12.5 mM α-D-lactose. The influence of protein folding on antibiofilm activity was also investigated. For this purpose, the lectin was tested after denaturation by heating 1 h at 100 °C.
Biofilm Mass Quantification
Quantification of biofilm mass was determined by crystal violet staining. After biofilm development, the content of each well was removed, and the wells were washed twice with 200 μL of 150 mM NaCl to remove weakly adherent cells. For fixation of biofilms, 200 μL of 99% methanol were added to each well. After 15 min, the methanol was removed, and the plates were allowed to dry at 25 °C. Then, 200 μL of crystal violet were added to each well. After 5 min, the excess of crystal violet was removed, and the plates were washed twice with water. Finally, 200 μL of acetic acid (33%, v/v) were added to wells to dissolve the crystal violet bound to the biofilm mass, and the absorbance was measured at 590 nm (OD590) using a microplate reader (SpectraMax® I3).
Quantification of Number of Biofilm Viable Cells
In order to determine the number of viable cells, the wells were washed twice with 150 mM NaCl, and then 200 μL of the same solution were added to each well containing biofilms. The plates were placed in an ultrasonic bath for 10 min to release biofilm-entrapped cells. Serial decimal dilutions from the obtained suspensions were plated on TSA to allow viability determination. The plates were then incubated for 24 h at 37 °C, and the number of colony forming units per milliliter (CFU/mL) was determined.
Statistical Analysis
Statistical analyzes were performed by GraphPad Prism® version 5.0 from Microsoft Windows®. The data from all assays were compared using one-way analysis of variance (ANOVA), with Bonferroni post hoc test. The data were considered significant when p < 0.05.
Results
Lectin Purification
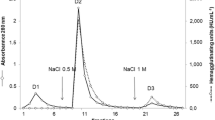
Crude extract of A. dactylomela eggs showed an evident and strong HA against human and rabbit erythrocytes. The HA induced by the treatment with extract was inhibited by the monosaccharide D-galactose and the disaccharide α-D-lactose. Affinity chromatography on HCl-activated Sepharose™ 4B was performed, and a novel lectin from sea hare eggs was successfully purified after elution with 0.2 M lactose (Fig. 1). In order to simplify the nomenclature, the new lectin was termed ADEL (derived from A. dactylomela eggs lectin). It was purified eight times and represents 81% of the total activity of the crude extract (Table 1).
ADEL purification. Affinity chromatography on HCl-activated Sepharose™ column (1.0 × 8.0 cm). The column was equilibrated and washed with TBS. Retained fractions were eluted with 0.2 M lactose in TBS. Approximately 20 mL of the crude extract were applied. Flow was 1 mL/min. SDS PAGE 15%. M molecular marker. ADEL in absence and presence of β-mercaptoethanol
Hemagglutinating Activity and Inhibition Assays
ADEL showed agglutinating activity in all tested erythrocytes in native and protease-treated forms. The lectin showed a slight preference for trypsin-treated rabbit erythrocytes (data not shown).
Several galactose-related carbohydrates showed inhibitory activity at distinct levels, with HA promoted by ADEL (Table 2). Galacturonic acid was the most potent inhibitor, followed by D-galactosamine and α-methyl-D-galactopyranoside. The minimum inhibitory concentrations (MIC) for these sugars were 0.2, 5, and 5 mM, respectively.
Biochemical Characterization
HA promoted by ADEL was optimal at pH 6 and 7. The lectin was stable in temperatures lower than 60 °C, and a complete loss of activity was achieved at 80 °C. EDTA and CaCl2 did not affect lectin activity (data not shown).
SDS-PAGE of ADEL resulted in 60 and 29 kDa bands in the absence and presence of β-mercaptoethanol, respectively, suggesting that ADEL is a dimeric protein formed by two identical polypeptide chains linked by a disulfide bond (Fig. 1). Native molecular mass estimated by SEC was 14 kDa, suggesting an anomalous behavior of the lectin in gel filtration (Fig. 2).
MALDI-TOF mass spectrometric analysis showed two distinct ions: [M + 1H]1+ at m/z 57,228 and [M + 2H]2+ at m/z 28,613, which agrees with the molecular mass estimated by SDS PAGE (data not shown).
The first 29 amino acid residues of ADEL were identified by amino acid sequencing. The N-terminal sequence 1DPDKCKTIRVESWSYKYAEKVVEDASYVL29 showed high similarity with a galacturonic acid-binding lectin from Aplysia kurodai.
Amino Acid Sequencing by MS/MS
The primary structure of ADEL was determined by overlapping of amino acid sequences of peptides sequenced by MS/MS deposited in Uniprot under accession number C0HK25. The amino acid sequence of ADEL consists of 217 residues, including 11 half-cystines involved in at least five intrachain disulfide bonds and one interchain disulfide bond (Fig. 3). According to the DiANNA 1.1 web server (Ferre and Clote 2005), cysteines are paired in the following form: 5Cys-Cys187, 42Cys-Cys68, 61Cys-Cys77, 114Cys-Cys135, and 142Cys-Cys206, whereas 172Cys is involved in maintenance of the dimer. The theoretical isoeletric point was 5.41.
Amino acid sequence of ADEL. Primary structure of ADEL was determined by overlapping of peptides obtained by digestions with trypsin (T), chymotrypsin (Q), and pepsin (P). Asterisks represent sequence microheterogeneites. Molecular mass determined by mass spectrometry are in parenthesis. Leucine and Isoleucine were assigned according with similarity between ADEL and AKL
Theoretical molecular mass for the amino acid sequence totaled 24,450 Da. However, this value is lower than the molecular mass determined by MALDI-TOF. This divergence is most likely a result of glycosylation in the polypeptide chain.
ADEL has four glycosylation sites, of which three contain attached glycans, as indicated by tryptic glycopeptides (Table 3). These peptides showed differences in their observed and calculated molecular mass. These differences were attributed to glycans with distinct masses.
To confirm the existence of glycans attached to the polypeptide backbone, a deglycosylation assay was performed. Peptides presented in Table 3 were already deglycosylated after treatment with PNGase F. For instance, peptide T-9 at m/z 1076.45 has a sequence of 118SFGFYNQNSTVWVNHGCR136, and its calculated molecular mass was 2171.95, whereas mass observed by MS was 3226.25. The difference between observed and calculated mass was 1054.3, corresponding to the glycan with composition HexNAc2Hex4. After PNGase F digestion, T-9 showed m/z 726.31 and observed molecular mass of 2172.9, indicating the release of the glycan (Fig. 4).
The RADAR tool (http://www.ebi.ac.uk/Tools/pfa/radar/) was employed to detect the presence of three tandem-repeat domains along the polypeptide chain, which share distinct levels of identity (Fig. 5). Domain A (2-70) has identity of 63 and 55% to domain B (74-144) and C (152-214), respectively, whereas domain B showed identity of 49% to domain C. Furthermore, ADEL presented microheterogeneities in three positions: 4K/M, 53D/A, and 199A/T.
ADEL showed high sequence similarity with AKL (A. kurodai eggs lectin, GenBank: AB968312.1) and with a hypothetical lectin isoform X1 (NCBI RefSeq XP_005101558.1) from Aplysia californica (Fig. 6).
Alignment of ADEL and other Aplysia eggs lectins. Amino acid sequence of ADEL was aligned with Aplysia kurodai eggs lectin (AKL) and a putative lectin from Aplysia californica (isoform X). Alignment realized by ESPript 3.0. Black and white boxes represent identical and non-identical amino acids, respectively. Consensus >50 represents amino acids present in at least 50% of the aligned sequences
Circular Dichroism
Spectra of native ADEL in far-UV exhibited minimum absorption at 215 nm and maximum at 202 and 229 nm, suggesting a predominance of β-conformation in its secondary structure (Fig. 7a).
CD spectra of ADEL. a Far-UV CD spectra (190–250 nm) of ADEL. The cuvette path length was 0.05 cm; protein concentration was 8 μM in PBS, pH 7. b Far-UV CD spectra of ADEL in different temperatures. c Near-UV CD spectra (250–330 nm) of ADEL with some ligands. The cuvette path length was 0.5 cm; protein concentration was 20 μM in PBS, pH 7
According to the CONTIN prediction method (Van Stokkum et al. 1990), available online as part of the server DICROWEB (http://dichroweb.cryst.bbk.ac.uk), the theoretical secondary structure of ADEL consisted of 4% α-helix, 44% β-sheet, 21% turn, and 32% coil.
Changes in the secondary structure of ADEL were examined by monitoring modifications in CD spectra as a function of temperature. Between 20 and 70 °C, no significant changes in secondary structure were observed, but after heating at 80 °C, some modifications occurred in maximum and minimal absorptions, indicating loss of secondary structure (Fig. 7b).
The spectra acquired in near-UV in the presence of some ligands revealed mild changes in tertiary structure of the lectin in comparison with native form (Fig. 7c). D-galactose (10 mM) caused a slight variation in tertiary structure of ADEL, while galacturonic acid (10 mM) exhibited strong effect on ADEL conformation.
Thermodynamics parameters of ADEL folding and unfolding in the presence and absence of carbohydrates are presented in Table 4. As demonstrated in the hemagglutinating assay, galacturonic acid is the most important ligand of ADEL; the interaction between galacturonic acid and ADEL shows Ka = 1.5 × 107 M−1. Furthermore, the presence of galacturonic acid increases ADEL Tm by 9 K in comparison to apo-lectin (data not shown).
ADEL Structural Prediction and Docking Calculation
Prior to molecular modeling, the ADEL sequence was used as input data for a PSI-BLAST (Ding et al. 2014) search against the protein data bank (PDB) to identify suitable template structures (http://blast.ncbi.nlm.nih.gov/Blast.cgi). No significant sequence similarity was found to any experimentally determined structure. Based on that, interplay between different modeling servers and software was performed to produce the most reliable structural prediction of ADEL.
The overall starting ADEL structure was obtained by I-TASSER. However, several poorly modeled patches were identified since no template structures were available on PDB. To overcome this issue, the structural predictions of each individual ADEL domain performed by QUARK were used to fill the structural gaps. The selected structures were used by MODELER to produce a complete ADEL model, with 99.1% of residues placed in allowed regions (90.7% on favorable regions) of the Ramachandran plot (Supplementary Fig. 1).
The obtained ADEL model (217 amino acids) possesses two independent domains connected by a short loop (Phe100-Val105), which were named domains A (Asp1-Leu99) and B (Glu106-Pro217) (Fig. 8a). Domain A is composed of a flat three-stranded and a curved five-stranded β-sheet, while domain B presents a flat three-stranded and a curved four-stranded β-sheet. According to I-TASSER, the crystal structure of the polymeric immunoglobulin receptor ectodomain from the rainbow trout Oncorhynchus mykiss (PDB 5F1S) is the structure most closely related to ADEL on PDB, despite its low sequence identity.
ADEL overall structure. a Monomer structure of ADEL represented as colored cartoon and lines with a transparent surface delineating the protein shape. The carbohydrate-binding site location is highlighted by a red dashed circle. b Carbohydrate-binding site representation, which shows the coordination of D-galacturonic acid by the main site residues (represented as green sticks)
The predicted binding site is located at the interface between domains A and B, mainly built by residues Lys6, Ile8, Tyr17, Glu106, Asp107, Gln109, and Arg207 (Fig. 8b).
Refined docking calculations resulted in an increasing order of theoretical binding energy among the analyzed carbohydrates, as follows: D-galactose < D-galactosamine < α-lactose < D-galacturonic acid (Table 5). Docking calculations also revealed a conserved binding pattern for the analyzed monosaccharides over the ADEL carbohydrate-binding site. It is possible to observe that the side chains of residues Lys6 and Ile8 act to stabilize the pyranosidic ring of the monosaccharides through hydrophobic stacking interactions. On the other hand, the hydroxyl groups of these ligands establish an intricate hydrogen bond pattern with the main chain carbonyl groups from residues Tyr17 and Asp107 and with the side chain polar groups from residues Glu106, Gln109, and Arg207 (Fig. 9 and Table 5).
Ligand coordination at the carbohydrate-binding site. a Coordination of D-galactose, represented as yellow sticks. b Coordination of D-galactosamine, represented as magenta sticks. c Coordination of D-galacturonic acid, represented as green sticks. d Coordination of α-lactose, represented as purple sticks. The protein backbone and the main amino acid residues involved in carbohydrate coordination by ADEL are represented as gray cartoon and sticks, respectively. Hydrogen bonds are represented as yellow dashes. A few interactions were omitted for the purpose of clarity (see Table 5)
In the case of α-lactose, the binding pattern seems to be slightly different. The pyranosidic rings from the D-galactose and D-glucose residues are stabilized not only through hydrophobic interactions with residues Lys6 and Ile8, but also with the side chain of residue Thr7. Further, the hydrogen-binding pattern also changes to accommodate this bulkier ligand (Fig. 9d and Table 5). It is worth mentioning that the ADEL carbohydrate-binding site is disposed in a groove between domains A and B surrounded by other solvent-exposed charged and hydrophobic amino acid residues that may comprise extended sites for the anchorage of complex ligands.
Agglutination and Antibacterial Activity
ADEL agglutinated S. aureus cells, but it was not able to agglutinate E. coli (Figure 10). Despite its agglutinating activity, ADEL was not able to inhibit the growth of either species (data not shown).
Effects of ADEL on Biofilm Formation
The effect of ADEL on the prevention of biofilm formation was assessed by two different assays: determination of biofilm mass and enumeration of biofilm viable cells. The treatment of S. aureus biofilms with ADEL reduced biomass by almost 40%. The effect was seen in all concentrations evaluated (Fig. 11a). Regarding the number of biofilm-entrapped viable cells, ADEL reduced S. aureus viability mainly at higher concentrations (250–62.5 μg/mL) (Fig. 11b). Interestingly, the biofilm formation of E. coli was not affected (Fig. 11).
The antibiofilm activity on S. aureus was abolished when ADEL was denatured by heating at 100 °C. On the other hand, treatment with α-D-lactose neither interfered with biofilm formation nor did it inhibit the antibiofilm activity of ADEL (data not shown).
Discussion
A new lectin from eggs of the sea hare A. dactylomela was successfully isolated and purified by affinity chromatography using Sepharose™ chromatography media. The use of Sepharose™ for purification of galactophilic lectins is a classical method, and several lectins have been successfully isolated by this procedure (Gasparini et al. 2008; Jimbo et al. 2000; Moura et al. 2006; Vasta and Marchalonis 1986). The activation of Sepharose™ by HCl constitutes an essential step to expose galactosides of the internal matrix, as long as these residues are available for specific lectin interaction.
The literature reports three lectins isolated from reproductive tissues of sea hares, which were isolated from Aplysia depilans (AGL) and A. kurodai (AKL-1 and AKL-2). AGL and AKL-1 are dimeric proteins with 30 kDa per subunit linked by disulfide bonds, as is ADEL. AKL-2 is also a dimeric protein, but with 16 kDa per subunit (Gilboa-Garber et al. 1985; Kawsar et al. 2009, 2011).
The literature shows that AKL-1, AKL-2, and AGL all have a strong affinity for galacturonic acid (Gilboa-Garber et al. 1985; Kawsar et al. 2009, 2011). ADEL also presents high specificity for the same carbohydrate. In fact, the specificity of lectins from sea hare eggs is a characteristic that deserves closer attention based on the fact that the species are spread far apart, but share the same sugar specificity. Thus, galacturonic acid should be considered as one of the preferred targets for the sea hare eggs lectins.
In addition to galacturonic acid, ADEL was also inhibited by D-galactose and its derivatives. ADEL showed slight preference by β-galactosides, thus suggesting the involvement of the hydroxyl at C-1 in the interaction between lectin and sugar. For instance, methyl-β-D-galactopyranoside and 4-nitrophenyl-β-D-galactopyranoside are two times more effective than methyl-α-D-galactopyranoside and 4-nitrophenyl-α-D-galactopyranoside, respectively. Moreover, β-lactose was more effective in inhibiting ADEL than α-lactose. The presence of substituents at C-1, such as methyl, phenyl, and nitrophenyl, increases inhibitory efficiency when compared to D-galactose. However, methyl-β-D-thiogalactopyranoside showed no inhibitory effect, indicating steric hindrance, most likely from the presence of a thiol group. Although C-1 seems to be an important recognition point, a similar grade of inhibition by melibiose (Galα1➔6Glc), α-D-lactose (Galβ1➔Glc), and raffinose (Galα1➔6Glcα1➔4Fru) indicates that C-1 cannot be considered as a key point in the recognition by ADEL.
Since D-glucose, L-fucose, and D-mannose are not recognized by ADEL, the orientation of hydroxyls at C-2 and C-4 seems to be more important than that at C-1. The axial orientation of C-2 and equatorial orientation of C-4 present in D-galactose seem to be essential for interaction. ADEL was inhibited to a greater degree by D-galactosamine compared to D-galactose, and it was not inhibited by GalNAc, indicating that a small uncharged substituent at C-2 (i.e., amine) increases affinity, whereas a charged substituent (i.e., acetyl) abolishes it.
The importance of C-5 was indicated by strong inhibition by the presence of a hydroxymethyl group in D-galacturonic acid when compared to the inhibitory effect produced by D-galactose, L-arabinose, and L-rhamnose, even though the first two sugars show the same orientation of hydroxyls at C-2, C-3, and C-4. The presence of the hydroxymethyl group strongly improves lectin-sugar binding since galacturonic acid is 6 × 105 times more effective than D-galactose.
The glycoprotein PSM has a terminal GalNAc and D-galactose as internal residue (Slomiany and Meyer 1972), and it exhibited inhibitory effect on ADEL. Like other sea hare lectins, ADEL was inhibited by small amounts of PSM (Kawsar et al. 2009). The presence of T-antigen (Galβ1➔3GalNAc) in PSM could account for the inhibitory effect based on the strong recognition of T-antigen by AGL (Gilboa-Garber and Sudakevitz 2001).
Consistent with data from the hemagglutination inhibition assay, CD spectrum of ADEL acquired in the near-UV spectral region revealed that the interaction with galacturonic acid plays an important role in ADEL conformation. D-galactose, D-galactosamine, and α-D-lactose also exerted some influence on the tertiary structure of ADEL, but to a lesser extent. These sugars showed distinct Ka values in the following order: galacturonic acid > D-galactosamine > D-galactose > α-D-lactose.
Curiously, the order of affinity seen in molecular docking was different in comparison to that observed in CD and HA inhibition. In the computational approach, galacturonic acid remains a sugar with higher binding energy, but α-D-lactose comes in second, followed by D-galactosamine and D-galactose.
In addition to clarifying the interactions between ADEL and sugars, CD measurement was also used to estimate secondary structure. According to the CD spectra, ADEL has a high content of β-conformations (sheets and turns), a characteristic of many lectins (Vasta et al. 2004). Indeed, the 3D model of ADEL is predominantly composed of β-structures, but interestingly, it presents a singular structure for lectins.
Animal lectins present a wide structural diversity. For instance, galectins have a typical jelly roll β-sandwich conformation (Vasta et al. 2004). The C-type lectin domain (CTLD) consists of a double loop stabilized by two disulfide bonds highly conserved in the base of the second loop (Zelensky and Gready 2005), while R-type lectins showed a β-trefoil (Uchida et al. 2004). However, none of these folds resembles the ADEL model. Obviously, for a more accurate comparison, the determination of three-dimensional structure of ADEL by experimental approaches is required.
ADEL, A. kurodai egg lectin and AGL are independent of divalent ions (Gilboa-Garber et al. 1985; Kawsar et al. 2009). AKL-1 and AGL are glycoproteins (Gilboa-Garber et al. 1985; Kawsar et al. 2009). In addition to evidence that AKL and AGL are glycoproteins, our study could provide accurate glycan structures of ADEL by MS analysis. ADEL is also a glycoprotein with three distinct glycans attached to three distinct N-glycosylation sites.
N-glycosylation is one of the most common post-translational modifications (PTMs) present in proteins, and several functions are attributed to its presence, including correct protein folding, protein localization, and regulation of protein degradation. Glycosylation events typically occur in the endoplasmatic reticulum (ER), and oligosaccharide maturation occurs in the Golgi apparatus (Ghazarian et al. 2011). According to the type of carbohydrate attached to polypeptide chain, N-oligosaccharides can be classified into three groups: high mannose, complex, and hybrid oligosaccharides (Kukuruzinska and Lennon 1998). Generally, all N-glycans share a common pentasaccharide core: Manα3(Manα6)Manβ4GlcNAcβ4GlcNAc. Our findings indicated that this precursor is also present in the ADEL structure.
In the first N-glycosylation site of ADEL (30NMT), an oligosaccharide of 1702.61 Da was found with mass corresponding to the pentasaccharide precursor (892 Da) with addition of five hexose residues (810 Da). In the second site (102NMT), molecular mass of the oligosaccharide ranges from 892.36 to 1216.45 Da, corresponding to the pentasaccharide precursor alone, and the precursor with two hexose residues (324 Da), respectively. Finally, the third site (126NST) has an oligosaccharide of 1054 Da that consists of the precursor and one hexose residue.
All oligosaccharides found in ADEL consist of a pentasaccharide precursor increased by some hexose residues. Among N-glycan groups, the only one that contains only hexoses, besides the precursor, is the high-mannose oligosaccharide.
Therefore, MS analysis strongly suggests that all oligosaccharides attached to the ADEL polypeptide chain are high-mannose-type oligosaccharides. To the best of our knowledge, this is the first time that glycan composition has been characterized for a lectin from opisthobranch (Gastropoda).
Although several efforts were made to remove oligosaccharides attached to ADEL, they were unsuccessful. Digestion of ADEL by PNGase F showed no effect, since no shift was observed in SDS PAGE and MS (data not shown). We speculate that the native folding of ADEL was maintained, even when using heat treatment and surfactant reagents during digestion. However, PNGase F could digest tryptic peptides of ADEL. All glycosylated peptides were digested by PNGase, indicating that no deoxyhexoses are attached to chitobiose precursors.
Besides glycosylation, specificity for carbohydrates, divalent ion independence, and quaternary arrangement, ADEL shares a high level of sequence similarity with AKL-1. ADEL showed 91 and 88% of identity and 97 and 93% of similarity with AKL-1 (GenBank: AB968312.1) and isoform X1 of A. californica (NCBI RefSeq XP_005101558.1), respectively.
Lectins isolated from the Aplysia genus showed no similarity to other known lectins, but the presence of a repeated domain in their structures is a common property of certain families of lectins. In particular, lectins that showed repeated domain in their primary structures and shared similarity of sequence with B-chain of Ricin were grouped in the R-type lectin family (Hirabayashi et al. 1998).
Lectins from sea hare eggs showed low similarity with Ricin, did not share conserved residues, and had no β-trefoil domain typical of R-type lectins, according to structural modeling. Thus, ADEL, AKL, and isoform X1 could not be grouped in this family, but could be grouped in a new family of lectins considering their similarities of sequence and biochemical characteristics. However, further experiments should be performed to better characterize these lectins.
ADEL was effective in agglutinating planktonic cells and inhibiting biofilm formation by S. aureus ATCC 25923. In fact, several galactophilic lectins can agglutinate bacterial cells (Carneiro et al. 2015; Melo et al. 2014; Moura et al. 2006), recognize glycans on the cell surface and distinguish Gram-positive from Gram-negative bacteria (Sun et al. 2007; Moura et al. 2006). The surface of Gram-positive bacteria is constituted by several structures containing carbohydrates, such as peptidoglycan, capsular polysaccharides, and teichoic acid (Weidenmaier and Peschel 2008), which may serve as possible binding sites for ADEL.
Lectins from marine organisms have been exploited for several biological applications, including antimicrobial activity (Moura et al. 2006; Roué et al. 2012; Cheung et al. 2015). Although some studies have focused on the antimicrobial potential of marine lectins, only a few are directly related to their effect on pathogenic biofilms. In the present work, we also evaluated the antibacterial potential of ADEL on planktonic growth and biofilm formation by S. aureus. ADEL does not inhibit bacterial growth, but rather inhibits biofilm formation. According to Liljemark et al. (1981), the formation of bacterial aggregates can cause a decrease in the number of adherent cells, which may explain why ADEL does not inhibit bacterial growth, but is, instead, effective on biofilm formation. Similar results were obtained by Vasconcelos et al. (2014), using plant and red algae lectins on Gram-positive and Gram-negative bacteria, as well as on yeasts biofilms.
In order to investigate the involvement of CRD in antibiofilm activity, the lectin was preincubated with α-D-lactose at MIC (12.5 mM). Interestingly, the activity was not abolished. This is not surprising since differences in affinity can be achieved between monosaccharides and structured glycans. Therefore, it is possible that ADEL has a higher affinity to complex glycans located on the S. aureus surface. Indeed, lectins can preferentially recognize carbohydrate complexes (Ghazarian et al. 2011). Furthermore, the inhibition of biofilm formation by ADEL is dependent on its structural integrity, in particular because denaturation by heating inhibited its antibiofilm activity.
Biofilms are described as structured communities of surface-associated microbial cells enclosed in a self-produced polymeric matrix (Flemming and Wingender 2010). Since biofilms are associated with increased resistance to conventional antimicrobial agents, ADEL could be considered as an alternative to preventing biofilm formation.
In addition, the antibiofilm activity exhibited by ADEL allows us to speculate about its physiological roles. For instance, Aplysia eggs are deposited on the surface of stones and algae where they remain for a few days until hatching. During incubation, eggs are exposed to several biological stresses, including bacterial infection. Therefore, the endogenous production of ADEL may be induced by the presence of pathogenic bacteria, and the lectin might act in the defense of eggs.
In conclusion, we herein report a novel type of lectin isolated from A. dactylomela, which is highly specific for galacturonic acid and has three high-mannose oligosaccharides attached to its polypeptide chain. Moreover, ADEL can inhibit biofilm formation by S. aureus. Thus, ADEL could be considered as an alternative to conventional antimicrobial therapy used to combat Staphylococcal biofilms.
References
Alpuche J, Pereyra A, Mendoza-Hernández G, Agundis C, Rosas C, Zenteno E (2010) Purification and partial characterization of an agglutinin from Octopus maya serum. Comp Biochem Physiol B 156:1–5
Bradford MM (1976) A rapid and sensitive method for quatitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem 72:248–534
Bulgakov A, Park KI, Choi KS, Lim HK, Cho M (2004) Purification and characterisation of a lectin isolated from the Manila clam Ruditapes philippinarum in Korea. Fish Shellfish Immunol 16:487–499
Carneiro RF, Melo AA, Almeida AS, Moura RM, Chaves RP, Sousa BL, Nascimento KS, Sampaio SS, Lima JP, Cavada BS, Nagano CS, Sampaio AH (2013) H-3, a new lectin from the marine sponge Haliclona caerulea: purification and mass spectrometric characterization. Int J Biochem Cell Biol 45:2864–2873
Carneiro RF, Teixeira CS, Melo AA, Almeida AS, Cavada BS, Sousa OV, Rocha BAM, Nagano CS, Sampaio AH (2015) L-rhamnose-binding lectin from eggs of the Echinometra lucunter: amino acid sequence and molecular modeling. Int J Biol Macromol 78:180–188
Chen J, Xiao S, Yu Z (2011) F-type lectin involved in defense against bacterial infection in the pearl oyster (Pinctada martensii). Fish Shellfish Immunol 30:750–754
Cheung RC, Wong JH, Pan W, Chan YS, Yin C, Dan X, Ng TB (2015) Marine lectins and their medicinal applications. Appl Microbiol Biotechnol 99:3755–3773
Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32:615–619
Ding S, Li Y, Shi Z, Yan S (2014) A protein structural classes prediction method based on predicted secondary structure and PSI-BLAST profile. Biochimie 97:60–65
Ferre F, Clote P (2005) DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res 33:230–232
Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633
Fujii Y, Kawsar SM, Matsumoto R, Yasumitsu H, Ishizaki N, Dogasaki C, Hosono M, Nitta K, Hamako J, Taei M, Ozeki Y (2011) A D-galactose-binding lectin purified from coronate moon turban, turbo (Lunella) coreensis, with a unique amino acid sequence and the ability to recognize lacto-series glycosphingolipids. Comp Biochem Physiol C 158:30–37
Fujii Y, Dohmae N, Takio K, Kawsar SM, Matsumoto R, Hasan I, Koide Y, Kanaly RA, Yasumitsu H, Ogawa Y, Sugawara S, Hosono M, Nitta K, Hamako J, Matsui T, Ozeki Y (2012) A lectin from the mussel Mytilus galloprovincialis has a highly novel primary structure and induces glycan-mediated cytotoxicity of globotriaosylceramide-expressing lymphoma cells. J Biol Chem 287:44772–44783
Gasparini F, Franchi N, Spolaore B, Ballarin L (2008) Novel rhamnose-binding lectins from the colonial ascidian Botryllus schlosseri. Dev Comp Immunol 32:1177–1191
Ghazarian H, Idoni B, Oppenheimer SB (2011) A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem 113:236–247
Gilboa-Garber N, Sudakevitz D (2001) Usage of Aplysia lectin interactions with T antigen and poly-N-acetyllactosamine for screening of E. coli strains which bear glycoforms cross-reacting with cancer-associated antigens. FEMS Immunol Med Microbiol 30:235–240
Gilboa-Garber N, Susswein AJ, Mizrahi L, Avichezer D (1985) Purification and characterization of the gonad lectin of Aplysia depilans. FEBS Lett 181:267–270
Greenfield NJ (2006) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc 1:2527–2535
Greenfield NJ (2007) Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat Protoc 1:2733–2741
He X, Zhang Y, Yu F, Yu Z (2011) A novel sialic acid binding lectin with anti-bacterial activity from the Hong Kong oyster (Crassostrea hongkongensis). Fish Shellfish Immunol 31:1247–1250
Hirabayashi J, Dutta SK, Kasai KI (1998) Novel galactose-binding proteins in Annelida: characterization of 29-kDa tandem repeat-type lectins from the earthworm Lumbricus terrestris. J Biol Chem 273:14450–14460
Ito S, Shimizu M, Nagatsuka M, Kitajima S, Honda M, Tsuchiya T, Kanzawa N (2011) High molecular weight lectin isolated from the mucus of the giant African snail Achatina fulica. Biosci Biotechnol Biochem 75:20–25
Jimbo M, Yanohara T, Koike K, Koike K, Sakai R, Muramoto K, Kamiya H (2000) The D-galactose-binding lectin of the octocoral Sinularia lochmodes: characterization and possible relationship to the symbiotic dinoflagellates. Comp Biochem Physiol B 125:227–236
Kawsar SMA, Matsumoto R, Fujii Y, Yasumitsu H, Dogasaki C, Hosono M, Nitta K, Hamako J, Matsui T, Kojima N, Ozeki Y (2009) Purification and biochemical characterization of a D-galactose binding lectin from Japanese sea hare (Aplysia kurodai) eggs. Biochemistry (Mosc) 74:709–716
Kawsar SMA, Matsumoto R, Fujii Y, Matsuoka H, Masuda N, Chihiro I, Yasumitsu H, Kanaly RA, Sugawara S, Hosono M, Nitta K, Ishizaki N, Dogasaki C, Hamako J, Matsui T, Ozeki Y (2011) Cytotoxicity and glycan-binding profile of a D-galactose-binding lectin from the eggs of a Japanese sea hare (Aplysia kurodai). Protein J 30:509–519
Kukuruzinska MA, Lennon K (1998) Protein N-glycosylation: molecular genetics and functional significance. Crit Rev Oral Biol Med 9:415–448
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227:680–683
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Liljemark WF, Bloomquist CG, Germaine GR (1981) Effect of bacterial aggregation on the adherence of oral streptococci to hydroxyapatite. Infect Immun 31:935–941
Matthews-Cascon H, Rocha-Barreira CA, Meirelles CAO (2011) Egg masses of some Brazilian mollusk. Expressão Gráfica e Editora, Fortaleza
Melo VMM, Duarte ABG, Carvalho AFFU, Siebra EA, Vasconcelos IM (2000) Purification of a novel antibacterial and haemagglutinating protein from the purple gland of the sea hare, Aplysia dactylomela rang, 1828. Toxicon 38:1415–1427
Melo AA, Carneiro RF, Melo WS, Moura RM, Silva GC, Sousa OV, Saboya JPS, Nascimento KS, Saker-Sampaio S, Nagano CS, Cavada BS, Sampaio AH (2014) HGA-2, a novel galactoside-binding lectin from the sea cucumber Holothuria grisea binds to bacterial cells. Int J Biol Macromol 64:435–442
Morris G, Huey H (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Moura RM, Queiroz AFS, Fook JMSLL, Dias ASF, Monteiro NKV, Ribeiro JKC, Moura GE, Macedo LL, Santos EA, Sales MP (2006) CvL, a lectin from the marine sponge Cliona varians: isolation, characterization and its effects on pathogenic bacteria and Leishmania promastigotes. Comp Biochem Physiol B 145:517–523
Mu C, Chen L, Zhao J, Wang C (2014) Molecular cloning and expression of a C-type lectin gene from Venerupis philippinarum. Mol Biol Rep 41:139–144
Naganuma T, Ogawa T, Hirabayashi J, Kasai K, Kamiya H, Muramoto K (2006) Isolation, characterization and molecular evolution of a novel pearl shell lectin from a marine bivalve, Pteria penguin. Mol Divers 10:607–618
Pales-Espinosa E, Perrigault M, Allam B (2010) Identification and molecular characterization of a mucosal lectin (MeML) from the blue mussel Mytilus edulis and its potential role in particle capture. Comp Biochem Physiol A Mol Integr Physiol 156:495–501
Roué M, Quévrain E, Domart-Coulon I, Bourguet-Kondracki ML (2012) Assessing calcareous sponges and their associated bacteria for the discovery of new bioactive natural products. Nat Prod Rep 29:739–751
Sampaio AH, Rogers DJ, Barwell CJ (1998) A galactose-specific lectin from the red marine alga Ptilota filicina. Phytochemistry 48:765–769
Shen M, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860
Slomiany DL, Meyer K (1972) Isolation and structural studies of sulphated glycoproteins of hog gastric mucin. J Biol Chem 247:5062–5070
Song X, Zhang H, Wang L, Zhao J, Mu C, Song L, Qiu L, Liu X (2011) A galectin with quadruple-domain from bay scallop Argopecten irradians is involved in innate immune response. Dev Comp Immunol 35:592–602
Sun J, Wang L, Wang B, Guo Z, Liu M, Jiang K, Luo Z (2007) Purification and characterisation of a natural lectin from the serum of the shrimp Litopenaeus vannamei. Fish Shellfish Immunol 23:292–299
Takahashi KG, Kuroda T, Muroga K (2008) Purification and antibacterial characterization of a novel isoform of the Manila clam lectin (MCL-4) from the plasma of the Manila clam, Ruditapes philippinarum. Comp Biochem Physiol B Biochem Mol Biol 150:45–52
Trott O, Olson AJ (2009) Software news and update Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Uchida T, Yamasaki T, Eto S, Sugawara H, Kurisu G, Nakagawa A, Kusunoki M, Hatakeyama T (2004) Crystal structure of the hemolytic lectin CEL-III isolated from the marine invertebrate Cucumaria echinata: implications of domain structure for its membrane pore-formation mechanism. J Biol Chem 135:37133–37141
Van Stokkum IHM, Spoelder HJW, Bloemendal M, Van Grondelle R, Groen FCA (1990) Estimation of protein secondary structure and error analysis from CD spectra. Anal Biochem 191:110–118
Vasconcelos MA, Arruda FV, Carneiro VA, Silva HC, Nascimento KS, Sampaio AH, Cavada BS, Teixeira EH, Henriques M, Pereira MO (2014) Effect of algae and plant lectins on planktonic growth and biofilm formation in clinically relevant bacteria and yeasts. Biomed Res Int. doi:10.1155/2014/365272
Vasta GR, Marchalonis JJ (1986) Galactosyl-binding lectins from the tunicate Didemnum candidum. Carbohydrate specificity and characterization of the combining site. J Biol Chem 261:9182–9186
Vasta GR, Ahmed H, Odom EW (2004) Structural and functional diversity of lectin repertoires in invertebrates, protochordates and ectothermic vertebrates. Curr Opin Struct Biol 14:617–630
Wang Y, Xiao Y, Suzek TO, Zhang J, Wang J, Bryant SH (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 37:1–11
Webb B, Sali A (2014) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. doi:10.1002/0471250953.bi0506s47
Weidenmaier C, Peschel A (2008) Teichoic acids and related cell-wall glycopolymers in gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287
Whitmore L, Wallace BA (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392–400
Wittmann V, Pieters RJ (2013) Bridging lectin binding sites by multivalent carbohydrates. Chem Soc Rev 42:4492–4503
Xu D, Zhang Y (2013) Ab initio structure prediction for Escherichia coli: towards genome-wide protein structure modeling and fold assignment. Sci Rep. doi:10.1038/srep01895
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:174–181
Yang J, Roy A, Zhang Y (2013) Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29:2588–2595
Zelensky AN, Gready JE (2005) The C-type lectin-like superfamily. FEBS J 272:6179–61217
Zhang J, Qiu R, Hu Y (2014) HdhCTL1 is a novel C-type lectin of abalone Haliotis discus hannai that agglutinates gram-negative bacterial pathogens. Fish Shellfish Immunol 41:466–472
Zheng P, Wang H, Zhao J, Song L, Qiu L, Dong C, Wang B, Gai Y, Mu C, Li C, Ni D, Xing K (2008) A lectin (CfLec-2) aggregating staphylococcus haemolyticus from scallop Chlamys farreri. Fish Shellfish Immunol 24:286–293
Acknowledgements
This work was supported by the Brazilian agencies CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FUNCAP (Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico), and FINEP (Financiadora de Estudos e Projetos). The authors thank CETENE for access to the mass spectrometer. The authors are especially grateful to Dr. Julia Campos for performing the MALDI-TOF experiments. The authors are grateful to Professor David Martin for helping in the English writing. A.H.S., C.S.N., and E.H.T. are senior investigators of CNPq.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Carneiro, R.F., Torres, R.C.F., Chaves, R.P. et al. Purification, Biochemical Characterization, and Amino Acid Sequence of a Novel Type of Lectin from Aplysia dactylomela Eggs with Antibacterial/Antibiofilm Potential. Mar Biotechnol 19, 49–64 (2017). https://doi.org/10.1007/s10126-017-9728-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-017-9728-x