Abstract
Gene targeting is a powerful tool for analyzing gene function. Recently, new technology for gene targeting using engineered zinc-finger nucleases (ZFNs) has been described in fish species. However, it has not yet been widely used for cold water and slow developing species, such as Salmonidae. Here, we present the results of successful ZFN-mediated disruption of the sex-determining gene sdY (sexually dimorphic on the Y chromosome) in rainbow trout (Oncorhynchus mykiss). Three pairs of ZFN mRNA targeted to different regions of the sdY gene were injected into fertilized rainbow trout eggs. Sperm from 1-year-old male founders (parental generation one or P1) carrying a ZFN-induced mutation in their germline were then used to produce F1 non-mosaic animals. In these F1 populations, we characterized 14 different mutations in the sdY gene, including one mutation leading to the deletion of leucine 43 (L43) and 13 mutations at other target sites that had different effects on the SdY protein, i.e., amino acid insertions, deletions, and frameshift mutations producing premature stop codons in the mRNA. The gonadal phenotype analysis of the F1-mutated animals revealed that the single L43 amino acid deletion did not lead to a male-to-female sex reversal, but all other mutations induced a clear ovarian phenotype. These results show that targeted gene disruption using ZFN is efficient in rainbow trout but depends on the ZFN design. We also characterized new sdY mutations resulting in male-to-female sex reversal, and we conclude that L43 seems dispensable for SdY function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The best way to understand the function of a gene of interest is to modulate its expression and characterize the resulting biological effects. Gene expression can be modulated at the transcript or protein level by using knockdown technologies, such as morpholinos (Heasman 2002) or RNAi (McManus and Sharp 2002; Dillin 2003; Tuschl 2002). However, a more effective method utilizes specific gene targeting, inducing a total loss of function by gene disruption (or knockout) or a gain of function by gene insertion (or knock-in). In the past, functional studies by gene targeting were restricted to mice, using embryonic stem cells modified through homologous recombination of DNA in the region of interest (for review, see Capecchi 2005). The availability of embryonic stem cells and knowledge on the full mouse genome allows specific gene targeting of essentially any gene of interest. However, even though genome-wide sequencing has been completed in additional species, embryonic stem cells have not yet been established for these species; thus, this powerful technology cannot yet be applied to domestic species, including cultured fish.
The rainbow trout (Oncorhynchus mykiss) is an important worldwide aquaculture species and is a model organism of considerable scientific importance (reviewed by Thorgaard et al. 2002). Many genomic resources are available for rainbow trout, including extensive classical expressed sequenced tag (EST) repertoires (Govoroun et al. 2006; Koop et al. 2008; Rexroad et al. 2003). With the development of next-generation sequencing (NGS) (Metzker, 2010), the identification of complex tissue-specific transcriptomes is now possible (Le Cam et al. 2012; Salem et al. 2010). This provides new opportunities to identify and characterize novel salmonid-specific genes. However, the functions of these genes cannot be deduced from their sequences, and modulation of gene expression is still needed.
Until recently, gene knockdown technology using antisense morpholino phosphorodiamidate oligonucleotides (AMOs) or siRNA was the only strategy available to investigate the in vivo functions of genes in rainbow trout (Boonanuntanasarn et al. 2002, 2003). Microinjection of AMOs into fertilized eggs was shown to efficiently block translation and mRNA splicing in rainbow trout, but the efficiency decreased drastically around the hatching stage. Therefore, the use of AMOs is restricted to the investigation of genes involved in early development, as their effect is limited in time (until hatching stage). Recently, zinc-finger nuclease (ZFN) technology has provided a powerful tool for editing the genomes of many plants and animals (for review, Carroll 2011), including some fish species such as zebrafish (Meng et al. 2008; Doyon et al. 2008), medaka (Ansai et al. 2012), and yellow catfish (Dong et al. 2011). By simple microinjection of ZFN mRNA into any fertilized egg, the ZFN introduces a specific and heritable double-strand break in the target sequence of the gene of interest (Bibikova et al. 2002).
Recently, we characterized a novel master sex-determining gene in rainbow trout. This gene, named sdY (sexually dimorphic on the Y chromosome), has been identified as a Y chromosome-specific gene that is conserved in many salmonids (Yano et al. 2012, 2013). Here, we report on additional technical details and mutations induced by different ZFNs that have been designed to target the sdY gene in rainbow trout. Because sdY is a Y chromosome gene, present as a single copy and only in the male genome, we can more easily detect and analyze an inactivation phenotype directly in the F1 generation. In addition, as sdY is known to be the master sex-determining gene in rainbow trout, loss of function of sdY using ZFNs should induce the feminization of genetically male (XY) individuals, and the characterization of this mutant phenotype can be easily determined by histological analysis of the gonads. These technical and biological advantages provide a favorable context to investigate the efficiency of ZFNs in a salmonid species. Therefore, in this study, we microinjected three different ZFNs targeted to two different regions of the sdY gene into fertilized rainbow trout eggs. The three ZFN pairs were initially designed to overcome any potential off-target effects of a single ZFN pair. Off-target effects were indeed a problem, as it was not possible to account for them in the in silico design owing to the absence of a publicly available full-genome sequence for rainbow trout.
Materials and Methods
Animals
Research involving animal experimentation conformed to the principles for the use and care of laboratory animals and complied with French and European regulations on animal welfare. Genetically all-male (XY) rainbow trout larvae were obtained from the Institut National de la Recherche Agronomique experimental fish farm (Drennec, France) as previously described (Guiguen et al. 1999). The eggs from the all-male populations were obtained by fertilization of normal eggs with sperm from YY males (Chevassus et al. 1988).
Zinc-Finger Nuclease Targeted Inactivation of sdY in Rainbow Trout
Targeted inactivation of sdY was generated using the CompoZr knockout ZFN technology according to the manufacturer's instructions (Sigma-Aldrich) and previously described protocols (Doyon et al. 2008; Amacher 2008). Three types of ZFN (ZFN1, ZFN2, and ZFN3) were designed to target two different sites in the sdY gene (Fig. 1). The ZFN target sequences were searched in all available rainbow trout transcript databases (Govoroun et al. 2006; Koop et al. 2008; Rexroad et al. 2003; Le Cam et al. 2012; Salem et al. 2010) to identify potential off-target sites. Fertilized eggs were activated in 1-mM reduced glutathione solution (pH 8.0) to prevent hardening of the chorion (Yoshizaki et al. 1991). A total of 100–500 ng/μl mRNA encoding the appropriate ZFN protein was injected into the blastodisc of all-male fertilized eggs as previously described (Yoshizaki et al. 1991).
ZFN target positions on the sequence of sdY exon 2. The primers used are shown in italics, the ZFN-targeted sequences are underlined, and the restriction enzymes sites are shown in bold type. To detect ZFN2- or ZFN3-induced mutations, we used the BslI restriction enzyme which cut the 343-bp sdY wild-type PCR product into three fragments of 96, 105, and 142 bp and the sdY PCR products containing a ZFN2- or ZFN3-induced mutation into two fragments of 201 and 142 bp. To detect ZFN1-induced mutations, we used the BtsCI restriction enzyme, which cut the 343-bp sdY wild-type PCR product into three fragments of 167, 114, and 62 bp and the sdY PCR products containing a ZFN1-induced mutation into two fragments of 281 and 62 bp. The positive PCR products showing only one restriction site (producing two digested PCR fragments) were verified by direct sequencing of the PCR product with the sdY E2Sa primer
Analysis of Germline Transmission
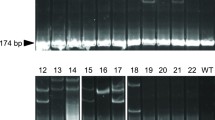
To characterize P1 animals carrying mutations transmitted to the germline, genomic DNA was extracted from the sperm of spermiating P1 males as previously described (Gharbi et al. 2006). Potential germline mutations were characterized using a CEL-I assay (SURVEYOR Mutation Detection Assay, Transgenomic), a technology based on a mismatch-specific endonuclease that cleaves all types of mismatches. This assay was performed on sdY PCR fragments containing the relevant ZFN cleavage site (Figs. 1 and 2). Sperm from the founders carrying a potential ZFN-induced mutation was then used to produce F1 animals. At 45 days postfertilization (dpf), the heads of 50–100 F1 embryos were collected for genomic DNA extraction. Genomic DNA from each F1 animal was extracted individually. A 5-fold dilution of the DNA samples was used for PCR with sdY primers (sdY Fw: 5′-CCCAGCACTGTTTTCTTGTCTC-3′ and sdY Rv: 5′-CTGTTGAAGAGCATCACAGGGTC-3′) to identify male individuals and to amplify the region surrounding the ZFN cleavage sites. The PCR conditions were as described (Yano et al. 2012a). To detect the mutations in male individuals, restriction enzyme digestion using BtsCI (New England Biolabs) for ZFN1 or BslI (New England Biolabs) for ZFN3 was performed on each of the F1 male animals. An absence of digestion at these sites indicated a mutation of this region by ZFN, and the PCR product of the mutated animals was directly sequenced.
A representative pattern of the characterization of mutations using the CEL-I assay in the germline (sperm) of P1 male founders. XY wild-type fish (XY control), ZFN1-mutated founder fish (ZFN1), and ZFN3-mutated founder fish (ZFN3) with (plus sign) or without (negative sign) addition of the CEL-I enzyme. Arrows show the cleaved DNA PCR fragments generated by the CEL-I enzyme, which creates double-strand breaks at nucleotide mismatches
Analysis of the Gonadal Phenotype of the ZFN-Mutated Fish
After 90 dpf, 288 F1 offspring were sacrificed from each P1 founder male with a positive germline mutation. A fin clip was taken for genomic DNA extraction, and a sample from the trunk was used for histological processing for each F1 animal. Genomic DNA from all of the F1 animals was extracted individually, and mutations were detected by restriction enzyme digestion as described above. PCR fragments from selected animals showing an absence of BslI or BtsCI sites were sequenced with the sdY Fw primer to confirm and characterize each mutation. All animals with a well-characterized mutation were fixed for at least 24 h in Bouin Holland fluid and embedded in paraffin. Each embedded sample was cut into 5-μm-thick sections and stained with Regaud's hematoxylin for histological analysis.
Results
Mutation Rate in ZFN-Induced Rainbow Trout
Three different pairs of ZFNs were designed (Fig. 1) to target two different regions of the sdY gene (ZFN2 and ZFN3 targeted the same region of the sdY gene). Potential off-target ZFN sequences were searched in rainbow trout databases, and no related sequences were found. The three pairs of ZFNs were injected into fertilized rainbow trout eggs. Macroscopic observation of the gonads of 1-year-old ZFN microinjected animals revealed that none of the P1 ZFN-mutated males had an ovarian phenotype. Therefore, sperm from the P1 founders was collected to determine germline mutation transmission. Using a CEL-I assay (Fig. 2), we detected 17 germline mutation transmissions from 70 ZFN1-injected mature fish (24 %), 5 germline mutation transmissions from 59 ZFN3-injected mature fish (8.5 %), and no germline mutation transmissions in 39 ZFN2-injected mature fish. Sperm from 9 founders carrying a ZFN1-induce nine mutations and two founders carrying a ZFN3-induced mutation were then used to produce F1 animals. The germline transmission rate was measured for each population by analyzing only male embryos (Table 1). The frequency of ZFN-mutated fish in the F1 populations ranged from 3 to 17 % (Table 1).
Sequence Analysis of ZFN Mutations
Fourteen different mutations in the sdY gene were detected from the nine mutant populations (eight ZFN1-mutated populations and one in the ZFN3-mutated population). Eight different nucleotide deletions, one insertion, and five combinations of nucleotide insertions and deletions were observed (Table 2, Fig. 3). Of the 14 different types of nucleotide mutation, six caused a frameshift leading to the generation of premature stop codons, four caused amino acid deletions (−1 to −5 AA), and the four remaining mutations were a combination of amino acid deletions, insertions, and substitutions (Fig. 3).
a Mutations in the sdY sequence induced by ZFN1. Nucleotide and derived protein sequences are shown for each mutation. A total of 13 different mutations in the sdY gene were detected in ZFN1 F1 animals; these included nucleotide deletion and insertion or deletion followed by insertion of nucleotides. b Mutated sdY sequence induced by ZFN3. Only one mutation was identified in the ZFN3 F1 animals. The asterisk represents a stop codon, resulting from a frameshift induced by the mutation in the sdY sequence. The numbers and frequencies of these mutations are listed in Table 2
Phenotypes of ZFN-Mutated Animals
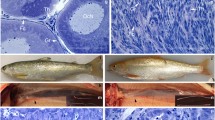
The gonad histology of the ZFN1-induced populations revealed a clear ovarian structure. This was characterized by the presence of the ovarian lamellae, which were delimited by connective tissue and contained meiotic oocytes (Fig. 4). In contrast, the ZFN3-mutated animals with a single amino acid deletion (the leucine at position 43) did not show any sex-reversal gonadal phenotype.
a Control XY male gonad at 100 days postfertilization (dpf), with germ cells shown by arrows and a characteristic dorsal blood vessel (BV) surrounded by a dotted line. b Control XX female gonad at 100 dpf showing a clear ovarian organization with ovarian lamellae (line) and meiotic germ cells (arrow head). ZFN1-mutated XY fish gonads at 100 dpf with a c Δ 14nt (shift/stop), d Δ 5nt;+2nt (−1 AA), and e Δ 5nt;+38nt (+11 AA) mutation presenting a male-to-female sex-reversal phenotype, with a clear ovarian organization showing ovarian lamellae (line) and meiotic germ cells (arrow head). f ZFN3-induced XY fish gonad with a Δ 3nt (−1 AA) mutation with no male-to-female sex-reversal phenotype
Discussion
sdY, the rainbow trout master sex-determining gene (Yano et al. 2012), was recently shown to be conserved in many salmonid species (Yano et al. 2013); however, it was not found in species outside the salmonid lineage. Because this gene is a salmonid innovation, it was necessary to analyze its function using a salmonid model such as rainbow trout. sdY is a Y chromosome gene that is present only in the male genome and as a single copy. Thus, the detection and characterization of an inactivation phenotype was possible in the F1 generation. In contrast, autosomal genes would require an additional generation (F2) to obtain a homozygous mutation, one that is present on both copies of the gene. The sdY gene was therefore a good target for investigating the possibility of generating loss-of-function mutations using the ZFN technology. We were able to create and characterize numerous mutations in the targeted gene, with identification of a clear associated phenotype for most of the mutations. Therefore, our results confirm the efficacy of ZFN technology in rainbow trout (Yano et al. 2012) and other fish species (Meng et al. 2008; Doyon et al. 2008; Ansai et al. 2012; Dong et al. 2011).
However, none of the P1 ZFN-mutated males displayed an ovarian phenotype, as would be expected for a loss-of-function mutation in a master sex-determining gene. This could be explained by possible cellular mosaicism of sdY mutations in the gonads of the P1 males. The mosaicism of the somatic cells in the mutated male gonads was not directly investigated in the present study. However, a high degree of mosaicism was detected during genotyping of the P1 animals before they reached maturity (data not shown). In addition, the low germline transmission rate (3–17 %), based on the mutation rate of F1 animals, also supports the hypothesis of an important mosaicism in the sdY loss-of-function mutations. This would suggest that even low expression of sdY can block the female differentiation pathway and/or induce the male differentiation pathway. These data fit with the idea that, as a master sex-determining gene acting at the top of the sex differentiation cascade, sdY has a very strong effect on the downstream pathway.
The percentage of male germline transmission detected in the P1 founders was highly variable, ranging from 0 % (ZFN2) to 25 % (ZFN1). The ZFN2 and ZFN3 pairs resulted in a lower percentage of germline transmission compared to ZFN1-injected populations. ZFN2 and ZFN3 pairs were designed to target the same region in the sdY sequence. Thus, the activity of these two pairs of ZFNs cannot be explained by the local structure of the genome that could interfere with nuclease recognition or function, such as chromatin architecture or DNA methylation status. The in vitro ZFN activities of these two different ZFN pairs, provided by the manufacturer and measured by the yeast MEL-1 reporter assay (Doyon et al. 2008), were very close (80.3 % for the ZFN2 pair and 74.3 % for the ZFN3 pair). The efficiency differences in vivo for these two constructs are most likely due to a suboptimal and unpredictable design, underlining the necessity of designing multiple ZFN pairs; this increases the likelihood of obtaining one pair with an efficient mutation rate in vivo.
In the F1 population, we observed low and variable germline transmission (3–17 %). This variability may be the consequence of the different concentrations of ZFN mRNA (100–500 ng/μl) that were injected into the P1 fertilized eggs. Initially, our study was designed to look at the effects of different ZFN mRNA concentrations. However, owing to space constraints of the experimental facilities, we were not able to keep the animals separated in different tanks; therefore, we could not differentiate between founders injected with a high or low concentration of ZFN mRNA. Space constraints are an important problem for large animals such as rainbow trout, especially as these experiments need to be conducted in well-controlled, recirculating water facilities to prevent escapees. However, our results demonstrated that, depending on the ZFN design, the efficiency can be good enough to produce germline mutations in 25 % of the microinjected fish. Combined with the fact that in some cases we were able to characterize multiple mutations in the same founder, an optimum strategy would be to design different ZFN pairs and keep only a limited number of founders to maturity.
We targeted two different positions of sdY with the ZFN pairs. This allowed us to investigate whether mutations in different parts of the protein could differentially affect the protein function and the resulting phenotype. For the ZFN3 pair, we observed only one mutation, which led to the deletion of leucine 43 (L43) and did not impact the sex phenotype. Many mutations were identified for the ZFN1 pair that targeted another region of the sdY gene; all of these mutations produced a clear sex-reversal phenotype. Among these ZFN1 mutations, we found one mutation leading to a single amino acid deletion of glycine 103 (G103). Interestingly, SdY G103 and L43 are both conserved, not only in all salmonid SdY proteins but also in the Irf9 interaction association domain (IAD) sequence that presents homologies with SdY (Yano et al. 2012, 2013). Although SdY is thought to be derived from an ancestral Irf9 duplication, the proteins are quite divergent in their overlapping domains; it is unknown whether the IAD domain is still functionally conserved in the SdY protein. However, the conservation of many amino acids between SdY and Irf9 suggests that some are likely to be crucial for the structure of both proteins. We show here that some of the conserved amino acids may not be as crucial for SdY function. For example, deletion of L43 does not affect the resulting sex phenotype. Comparison of the SdY sequences within salmonids shows that the region containing the ZFN1 target site is much more conserved (11/12 amino acids, i.e., 99.6 % identity) than the region containing the ZFN3 target site (8/12 identical amino acids, i.e., 66.6 % identity). These data suggest that the structure of the protein around the ZFN1-targeted region is more constrained than the region targeted by ZFN3. This hypothesis would explain why all the mutations characterized for the ZFN1 pair produced a clear sex-reversal phenotype, while the single mutation characterized with the ZFN3 pair did not produce a sex-reversal phenotype.
Unexpected off-target effects are a significant concern in targeted gene modification technologies. We did not identify any conservation of the ZFN-targeted sequences in the publicly available rainbow trout transcriptomes (Govoroun et al. 2006; Koop et al. 2008; Rexroad et al. 2003; Le Cam et al. 2012; Salem et al. 2010). This suggested that there were no potential off-target sites for the ZFN pairs that we used, at least in the protein-coding regions of the trout genome. The fact that we did not observe any unexpected phenotypic abnormalities in the mutated ZFN embryos (both at F1 and F2) also supports the absence of major off-target events. Genome editing with ZFNs has been reported in many species: plants (Shukla et al. 2009), Drosophila (Beumer et al. 2006), Xenopus (Young et al. 2011; Nakajima et al. 2012), fish (Meng et al. 2008; Doyon et al. 2008; Ansai et al. 2012; Dong et al. 2011), and many mammalian species (Carroll 2011), including domestic animals such as pigs (Watanabe et al. 2010; Whyte et al. 2011) and cattle (Yu et al. 2011). Accordingly, ZFN technology has high potential to produce gene knockouts in non-model species including salmonids. Our result shows that ZFN works in a cold-water species in which embryonic development is slow, and the generation times are extremely long. The ZFN approach provides the opportunity to characterize the function of novel salmonid-specific genes that are difficult to analyze using AMOs or siRNA.
References
Amacher SL (2008) Emerging gene knockout technology in zebrafish: zinc-finger nucleases. Brief Funct Genomic Proteomic 7:460–464
Ansai S, Ochiai H, Kanie Y, Kamei Y, Gou Y, Kitano T, Yamamoto T, Kinoshita M (2012) Targeted disruption of exogenous EGFP gene in medaka using zinc-finger nucleases. Dev Growth Differ 54:546–556
Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D (2006) Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics 172:2391–2403
Bibikova M, Golic M, Golic KG, Carroll D (2002) Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161:1169–1175
Boonanuntanasarn S, Yoshizaki G, Takeuchi T (2003) Specific gene silencing using small interfering RNAs in fish embryos. Biochem Biophys Res Commun 310:1089–1095
Boonanuntanasarn S, Yoshizaki G, Takeuchi Y et al (2002) Gene knock-down in rainbow trout embryos using antisense morpholino phosphorodiamidate oligonucleotides. Mar Biotechnol (NY) 4:256–266
Le Cam A, Bobe J, Bouchez O, Cabau C, Kah O, Klopp C, Lareyre JJ, Le Guen I, Lluch J, Montfort J, Moreews F, Nicol B, Prunet P, Rescan PY, Servili A, Guiguen Y (2012) Characterization of rainbow trout gonad, brain and gill deep cDNA repertoires using a Roche 454-Titanium sequencing approach. Genetics 500:32–39
Capecchi MR (2005) Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 6:507–512
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Chevassus B, Devaux A, Chourrout D, Jalabert B (1988) Production of YY rainbow trout males by self-fertilization of induced hermaphrodites. J Hered 79:89–92
Dillin A (2003) The specifics of small interfering RNA specificity. Proc Natl Acad Sci U S A 100:6289–6291
Dong Z, Ge J, Li K, Xu Z, Liang D, Li J, Li J, Jia W, Li Y, Dong X, Cao S, Wang X, Pan J, Zhao Q (2011) Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc finger nucleases. PloS one 6:e28897
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26:702–708
Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, Høyheim B, Taggart JB, Cairney M, Powell R, Krieg F, Okamoto N, Ferguson MM, Holm LE, Guyomard R (2006) A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172:2405–2419
Govoroun M, Le Gac F, Guiguen Y (2006) Generation of a large scale repertoire of expressed sequence tags (ESTs) from normalised rainbow trout cDNA libraries. BMC genomics 7:196
Guiguen Y, Baroiller JF, Ricordel MJ, Iseki K, Mcmeel OM, Martin SA, Fostier A (1999) Involvement of estrogens in the process of sex differentiation in two fish species: the rainbow trout (Oncorhynchus mykiss) and a tilapia (Oreochromis niloticus). Mol Reprod Dev 54:154–162
Heasman J (2002) Morpholino oligos: making sense of antisense? Dev Biol 243:209–214
Koop BF, von Schalburg KR, Leong J, Walker N, Lieph R, Cooper GA, Robb A, Beetz-Sargent M, Holt RA, Moore R, Brahmbhatt S, Rosner J, Rexroad CE 3rd, McGowan CR, Davidson WS (2008) A salmonid EST genomic study: genes, duplications, phylogeny and microarrays. BMC genomics 9:545
McManus MT, Sharp PA (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev Genet 3:737–747
Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA (2008) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26:695–701
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31–46
Nakajima K, Nakajima T, Takase M, Yaoita Y (2012) Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev Growth Differ 54:777–784
Rexroad CE, Lee Y, Keele JW, Karamycheva S, Brown G, Koop B, Gahr SA, Palti Y, Quackenbush J (2003) Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res 102:347–354
Salem M, Rexroad CE, Wang J, Thorgaard GH, Yao J (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11:564
Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459:437–441
Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ, Walsh PJ, Vijayan MM, Devlin RH, Hardy RW, Overturf KE, Young WP, Robison BD, Rexroad C, Palti Y (2002) Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B Biochem Mol Biol 133:609–646
Tuschl T (2002) Expanding small RNA interference. Nature Biotechnol 20:446–448
Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H (2010) Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun 402:14–18
Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, Laughlin MH, Prather RS (2011) Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev 78:2
Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, Bouchez O, Fostier A, Guiguen Y (2012) An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol 22:1423–1428
Yano A, Nicol B, Jouanno E, Quillet E, Fostier A, Guyomard R, Guiguen Y (2013) The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. doi:10.1111/eva.12032
Yoshizaki G, Oshiro T, Takashima F (1991) Introduction of carp α-globin gene into rainbow trout. Nippon Suisan Gakk 57:819–824
Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B (2011) Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A 108:7052–7057
Yu S, Luo J, Song Z, Ding F, Dai Y, Li N (2011) Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 21:1638–1640
Acknowledgments
We thank the LPGP experimental facility staff for their help in maintaining our ZFN rainbow trout. This work was supported by funds from INRA, ANR (SVSE 7 2011, project SDS), and the European Commission Seventh Framework Program (222719–LIFECYCLE). A.Y. and B.N were supported by a postdoctoral fellowship from the INRA GA and PHASE departments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yano, A., Nicol, B., Jouanno, E. et al. Heritable Targeted Inactivation of the Rainbow Trout (Oncorhynchus mykiss) Master Sex-Determining Gene Using Zinc-Finger Nucleases. Mar Biotechnol 16, 243–250 (2014). https://doi.org/10.1007/s10126-013-9546-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-013-9546-8