Abstract
Chlamys farreri is a significant species in aquaculture and fishery in East Asia. A deep understanding of its shell formation by studying the transcriptome of the mantle, a key organ in shell formation, could provide important guidance for its culture. Thus, we sequenced and analyzed the mantle transcriptome of C. farreri. The 77,975 unigenes were generated after Illumina sequencing and de novo assembly. The unigenes were annotated using authoritative databases (non-redundant (NR), COG, Gene Ontology (GO), and KEGG) to obtain functional information. BLASTX alignment was performed between unigenes and reported proteins related to biomineralization. The results identified 53 homologous genes representing 17 matrix proteins, most of which are involved in calcite formation, and 171 homologies with 26 proteins related to general processes of biomineralization. The discovery and unusually high expression of MSP-1 suggested its importance in scallops. Homologous unigenes with aragonite-formation-related matrix proteins were much fewer compared with those related to calcite formation. The results implied that, in C. farreri, the number and proportion of matrix proteins related to aragonite formation is much lower than those related to calcite formation, which was consistent with the proportions of aragonite and calcite in C. farreri shells. Thus, the formation of different polymorphs of calcium carbonate (calcite and aragonite) in molluskan shells is regulated by different groups of proteins. Moreover, 17 candidate unigenes, which are probably involved in biomineralization, were predicted by screening for gene products with secreted domains and tandem-arranged repeat units. Our results contribute to the understanding of biomineralization processes and the evolution of shell formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biomineralization, which refers to the dynamic physiological process by which a living organism elaborates a mineralized structure, is widely distributed in the metazoan taxa. The products of biomineralization include over 60 different kinds of minerals, like calcite and aragonite (Lowenstam 1981). The minerals synthesized by biomineralization are major materials used to build various organs, such as bones, teeth, and mollusk shells, which play significant roles in many species. The understanding of biomineralization benefits researches of many fields, such as medicine, and development of biomaterials and aquaculture and also has great commercial value. In biomineralization research, shell formation in mollusks is a hot topic. According to previous studies, mollusk shells usually comprise calcium carbonate and organic macromolecules (proteins, polysaccharides, and lipid) (Lowenstam and Weiner 1989). The organic macromolecules in mollusk shells, although only representing a minor proportion (around 5 %) of the total weight of the shell, have vital functions in the crystallization process of calcium carbonate (Belcher et al. 1996). Among these macromolecules, shell matrix proteins have received the most attention from researchers. Shell matrix proteins are considered to play essential roles in shell formation processes, such as the nucleation, growth, and regulation of calcium carbonate crystals. In the past decades, some of the matrix proteins have been separated and identified; however, it is believed that there are further undiscovered matrix proteins, and the exact functions, interactions, and regulation of the proteins remain undetermined (Zhang and Zhang 2006).
There are two major polymorphs of calcium carbonate in mollusks: calcite and aragonite. Shells are usually composed of calcite, aragonite, or both. Most shells are spatially separated into different layers, which are differentiated by their ultrastructural motifs. Among the various bivalve shell ultrastructures, some of them are made of calcite, like simple prismatic structure and foliated structure, and some of them are composed of aragonite, like nacreous structure and crossed lamellar structure (Taylor 1973; Lowenstam and Weiner 1989). In the reported matrix proteins, Nacrein (Miyamoto et al. 1996), N16/pearlin (Samata et al. 1999), and N19 (Yano et al. 2007) have been proved to be involved in the formation of aragonite. On the other hand, MSI31 (Sudo et al. 1997), MSI7 (Zhang et al. 2003), MSP-1 (Sarashina and Endo 1998; Sarashina and Endo 2001), Prismalin-14 (Suzuki et al. 2004), Aspein (Tsukamoto et al. 2004), the KRMP family (Zhang et al. 2006), Prisilkin-39 (Kong et al. 2009), and some other proteins are related to the formation of calcite.
Shell formation is a sophisticated biological process, which requires the participation and cooperation of many molluskan organs, of which mantle is the most important. Most matrix proteins, including those mentioned above, are specifically expressed in the molluskan mantle (Zhang and Zhang 2006). Thus, the mantle is a major subject of biomineralization research.
The transcriptome is the complete set of transcripts in a cell, knowledge of which is essential for exploring the functional elements of the genome and revealing the molecular constituents of cells and tissues (Wang et al. 2009). Compared with traditional research methods, deep RNA sequencing (RNA-seq) has an apparent advantage because of its capacity to produce large amounts of precise data in a short time (Metzker 2010). With the help of this approach, the qualitative and quantitative research of transcripts becomes available. This allows researchers to identify the expression conditions of certain genes and compare gene expression between different tissues or under different biological conditions. This information contributes greatly to the investigation of cellular functions (Mutz et al. 2012). Transcriptome-related analysis and experiments carried out earlier in other species also revealed a large amount of information about biomineralization and greatly promoted biomineralization research in the corresponding mollusks (Fang et al. 2011; Zhao et al. 2012; Shi et al. 2013; Werner et al. 2013).
Chlamys farreri, also known as the Zhikong scallop, is a Pacific Asian subtropical species (Shumway 1991). The scallop is distributed along the coasts of North China, Korea, Japan, and eastern Russia. C. farreri is a popular seafood in East Asian countries because of its large and edible adductor muscle, which makes it a significant species in aquaculture and fishery (Zhang et al. 2011). Thus, C. farreri have been an important research subject for decades; however, most of the previous studies emphasized aquaculture (Xiao et al. 2005) or immunology (Miao et al. 2011). Considering the positive correlation between the growth of the scallop and its shell formation (Shumway 1991), we believe that our research in shell formation of C. farreri can provide useful information for its culture. Moreover, since the shell structure of C. farreri differs from traditionary objects of biomineralization study, such as the Pinctada fucata, the exploration of shell formation in C. farreri could also improve the diversity of biomineralization research and assist our understanding of the sophisticated processes of biomineralization and the evolution of shell formation in this species.
Limited studies have been carried out in C. farreri regarding biomineralization, and transcriptome data are capable of providing massive amounts of information; therefore, sequencing and analysis of the C. farreri mantle transcriptome could be a promising start for research into the biomineralization process in this mollusk.
Materials and Methods
Total RNA Extraction
Adult individuals of C. farreri were collected and transported from Taiping Corner (Yellow Sea, Qingdao, China). The scallops were cultured in aerated artificial sea water (Sude instant sea salt, 3 %) for 3 days before the growing edge of the mantle tissue of the scallops was dissected and powdered in liquid nitrogen immediately. Total RNA was extracted following the instructions of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The integrity and quality of the total RNA were assessed by gel electrophoresis and measurement of the OD260 and OD260/OD280, using an Ultrospec 3000 UV/visible spectrophotometer (Amersham, Piscataway, NJ, USA).
cDNA Library Preparation and Illumina Sequencing
After the extraction of total RNA, magnetic beads coated with Oligo (dT) were used to isolate mRNA from total RNA. The mRNA was then mixed with fragmentation buffer to fragment the mRNA into short fragments. These mRNA fragments were used as templates to synthesize cDNA, which was later purified and dissolved in EB buffer for end repair and single nucleotide A (adenine) addition. Adapters were then added to the short fragments, and suitable fragments were selected as templates for polymerase chain reaction (PCR) amplification. The quantity and quality of the cDNA library were using the assistant software of the Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System. After the library was successfully established, it was sequenced using an Illumina HiSeq™ 2000.
De Novo Assembly and Sequence Annotation
After the acquisition of raw reads, reads with adaptors, and those with a percentage of unknown nucleotides greater than 5 % or a percentage of low-quality bases (base quality ≤10) more than 20 % were discarded to obtain clean reads. Transcriptome de novo assembly was carried out using the short reads assembly program, Trinity (Grabherr et al. 2011). After gene family clustering, the unigenes were divided into clusters and singletons. To decide the sequence direction and predict the protein coding regions of the unigenes, BLASTX alignments (e-value < 10−5) between the unigenes and protein databases (in order of priority: NR, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and the cluster of orthologous groups of proteins (COG)) were performed. If a unigene could not be aligned to any of the databases, we used ESTScan (Iseli et al. 1999) to decide its sequence direction and possible protein coding region. The protein coding region sequences were then translated into amino sequences using the standard codon table.
To obtain annotations, the unigenes were aligned by BLASTX to protein databases NR, Swiss-Prot, KEGG, and COG (e-value < 10−5), and aligned by BLASTN to nucleotide databases Nt (e-value < 10−5). Proteins with the highest sequence similarity with the given unigenes, along with their protein functional annotations, were retrieved. With NR annotation, the Blast2GO program (Conesa et al. 2005) was used to obtain GO annotations of the unigenes. After obtaining GO annotations for each unigene, we used WEGO software (Ye et al. 2006) to perform GO functional classification for all unigenes and determine the distribution of gene functions of C. farreri at the macro level.
Identification of Genes Involved in Biomineralization
The identification of homologous unigenes with biomineralization-related proteins was carried out by searching key words of reported proteins in the BLASTX alignment results with the databases mentioned above. Query proteins with negative feedback are not listed in Tables 3 and 4.
Gene Screening
For secreted protein screening, the unigenes in the S category (function unknown) of COG annotation and the ones not annotated to any databases were selected, among which the unigenes with a translated amino acid sequence shorter than 50 amino acid residues or with no stop codon were discarded. The translated amino acid sequences of the remaining unigenes were searched for signal peptides using SignalP v4.1 (http://www.cbs.dtu.dk/services/SignalP/) (D-cutoff values = sensitive) (Nielsen et al. 1997; Petersen et al. 2011). The unigenes predicted to contain signal peptides were then searched with TargetP v1.1 (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al. 2000) to rule out proteins targeted to mitochondria or other organelles. Transmembrane proteins were predicted with the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and then removed. Glycosyl phosphatidyl inositol (GPI)-anchored proteins were rejected using the GPI modification predictor (http://mendel.imp.ac.at/sat/gpi/gpi_server.html) (Eisenhaber et al. 1998; Sunyaev et al. 1999; Eisenhaber et al. 1999, 2000). To identify proteins with tandemly arranged repeat units, the data set was also submitted to XSTREAM (http://jimcooperlab.mcdb.ucsb.edu/xstream/) (Newman and Cooper 2007).
Results
Sequence Analysis and De Novo Assembly
After Illumina sequencing, 59,918,916 raw reads were generated (Table 1). Then reads with adaptors, a percentage of unknown nucleotides greater than 5 % or a percentage of low-quality bases (base quality ≤10) more than 20 % were filtered out, leaving 55,122,820 clean reads, which were subjected to de novo assembly. The clean reads contained 4,961,053,800 nucleotides, with an average length of 90 nucleotides. The values of the Q20 percentage and N percentage were 97.26 % and 0.01 %, respectively. In addition, the GC percentage of the clean reads was 44.13 %.
The genome map of C. farreri has not been revealed yet; therefore, we chose de novo assembly to assemble the sequencing data (Table 2). As a result, 186,629 contigs were generated, with a total length of 46,529,756 nucleotides and an average length of 249 nucleotides. These contigs were subsequently assembled into 77,975 unigenes. The total length of the unigenes was 41,985,113 nucleotides, and the mean length was 538 nucleotides. Among the unigenes, 14,287 were distinct clusters, and the other 63,688 were singletons.
Functional Annotation of the Unigenes
To obtain possible functional information of the unigenes, they were aligned by BLASTX to the protein databases NR, Swiss-Prot, KEGG, and COG (e-value < 10−5) and aligned by BLASTN to the nucleotide database Nt (e-value < 10−5). Proteins with the highest sequence similarity with the given unigenes were retrieved, along with their protein functional annotations.
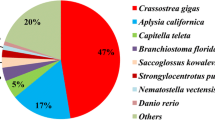
In the alignment with NCBI NR protein database, 31,474 unigenes were annotated, which accounted for 40.36 % of the assembled unigenes (Fig. 1). Among these successfully annotated unigenes, 30.8 % of them had strong homology with the proteins aligned (e-value less than 1.0E-45). The similarity distribution illustrated that 37 % of the sequences had a similarity higher than 60 %. As for the species distribution, most of the annotated unigenes were annotated to proteins from Crassostrea gigas (67.3 %).
Statistics of annotation results according to the NR database. a E-value distribution of unigenes annotated to the NR database; the cut-off e-value was 1.0E-5. b Similarity distribution between the unigenes and their homologs in the NR database. c Species distribution of the homologous genes of the annotated unigenes
To obtain a deeper understanding of the functions of the unigenes, BLASTX alignment was performed between the unigenes and the COG database (Fig. 2). The 8,777 unigenes were classified into 25 functional categories. Among those categories, the R category (general function prediction only) contained the largest number of unigenes (3,742; 42.63 %). Other categories we were interested in were the P category (inorganic ion transport and metabolism) and the S category (function unknown), because the genes related to biomineralization were most likely sorted into these two categories. The number of unigenes annotated into the P category and S category were 433 (4.93 %) and 1,376 (15.68 %), respectively.
The 13,811 unigenes were annotated by GO analysis (Fig. 3). Among them, 56,113 unigenes were annotated to the ontology of biological process, 35,276 to cellular component, and 16,578 to molecular function. The excess numbers occurred because one unigene might be aligned to different sub-categories. In the 61 sub-categories identified, “cellular process” under the ontology of biological process contained the maximum number of unigenes (9,136; 66.15 %).
The KEGG database was also searched to obtain information about the biological pathways operating in C. farreri. According to the alignment results, 20,063 unigenes were annotated into 259 different pathways in the KEGG database. Among them, metabolic pathways contained the largest number of unigenes (2,478; 12.35 %). Another pathway that aroused our interest was the calcium signaling pathway, which probably plays a significant role in the biomineralization process. Four hundred ten (2.04 %) unigenes were found to be involved in this signaling pathway, including unigenes homologous with calmodulin, a key element in shell formation related to calcium metabolism.
Screening of Genes Involved in Biomineralization
To obtain more information about the shell formation process of C. farreri, we searched the transcriptome data set for typical matrix proteins that had been reported and also proteins that had been proved to assist the shell formation process.
After the alignment with known matrix protein genes, 53 homologous unigenes of 17 matrix proteins, such as MSP-1, Shematrin-2, Aspein, and Prisilkin-39, were discovered in the transcriptome of C. farreri (Table 3). Previous studies linked most of them to the formation of calcite. MSP-1 is an unusually acidic protein enriched in Ser, Asp, and Gly and was isolated from the foliated calcite shell layer of another scallop, Patinopecten yessoensis (Sarashina and Endo 1998). In addition, the fragments per kilobase per million fragments (FPKM) values of MSP-1 homologies were remarkably high (Table 3), which indicated that the homologous unigenes are highly expressed in the C. farreri mantle. This high expression level indicates strongly that MSP-1 may play crucial roles in the shell formation of C. farreri. We also searched for genes related to aragonite formation, and only homologies with N66, Lustrin A, and Perlucin were found in the C. farreri mantle transcriptome.
In the alignment with proteins involved in the general processes of biomineralization, such as calcium signaling and metabolism, 171 unigenes homologous with 26 proteins were identified (Table 4). The existence of chitin synthase, carbonic anhydrase, alkaline phosphatase, G protein, calcineurin B, calmodulin-like protein (CaLP), and some other proteins reported to be related to shell formation was confirmed.
Prediction of Genes Potentially Related to Biomineralization
To identify further genes that might be involved in biomineralization, we used several bioinformatic methods to select and predict possible biomineralization-related genes. Since most of the matrix proteins participating in shell formation are secreted proteins, screening for proteins with signal peptides is the first step of the prediction. Based on the hypothesis that unidentified matrix protein genes were most likely annotated into the S category (function unknown) of the COG database or annotated into none of the databases, we searched for signal peptides in all proteins representing the unigenes in that category using SignalP. Two hundred seventeen unigenes were predicted to have a signal peptide. TargetP, the TMHMM Server, and GPI modification predictor were then used to rule out proteins targeted to mitochondria or any other organelles, and those with transmembrane domains or GPI-anchors. After filtering using these programs, 125 unigenes remained.
Another important feature of biomineralization-related proteins is that they are organized in different functional domains, most of which comprise tandemly arranged repeat units (Jackson et al. 2010; Fang et al. 2011). To identify proteins with tandemly arranged repeat units, the 125 unigenes obtained above were also screened using XSTREAM. Seventeen of them contained tandemly arranged repeat units (Fig. 4). The details of these unigenes and their tandemly arranged repeat units are shown in Supplementary Table S1. These 17 unigenes might be important candidates for subsequent study of the shell formation of C. farreri.
Schematic of unigenes predicted to be potential participants in shell formation. The products of unigenes were first screened by SignalP, TargetP, TMHMM, and GPI modification predictors for signal peptides. The outcome was then examined by XSTREAM for tandemly arranged repeat units. The retrieved unigenes and the description of their tandemly arranged repeat units are shown. The repeat units are represented by colored motifs. Each color represents a certain kind of repeat unit, and the identities of the units are marked by the number on top of each motif (red font). The length of the bars does not reflect the number of amino acids of the unigene products
Discussion
In our study, we extracted total RNA from the mantle tissue of the adult individuals of C. farreri. After cDNA library construction and Illumina sequencing, 59,918,916 raw reads were generated, of which 55,122,820 were clean reads. The Q20 percentage represents the proportion of nucleotides with quality values larger than 20 in reads. The N percentage equals the proportion of unknown nucleotides in clean reads. Q20 percentage and N percentage are two significant criteria to evaluate the quality of sequencing data. The value of the Q20 percentage (97.26 %) and the N percentage (0.01 %) suggested that the quality of our sequencing was acceptable. De novo assembly was carried out because the genome map has not yet been completed. The 77,975 unigenes were produced with an average length of 538 nucleotides. Thus, the assembly provided abundant materials for subsequent annotation and analysis.
The annotation to NCBI NR protein database revealed that most of the annotated C. farreri unigenes were homologous with genes of C. gigas (67.3 %). The result may be because C. gigas is one of the few mollusks whose genomes have been sequenced, and the details of numerous genes of C. gigas have been released (Zhang et al. 2012). Altough the evolutionary relationship between C. gigas and C. farreri is not close, the strong similarity revealed in our study still implied that studies of C. gigas can be reliable references for the study of C. farreri.
The 8,777 unigenes were annotated to the COG database, among which the unigenes in the P category (inorganic ion transport and metabolism) and the S category (function unknown) attracted most of our attention in subsequent analysis. The transportation and metabolism of calcium ions makes a substantial contribution to the biomineralization process (Belcher et al. 1996); thus, some unigenes in the P category were found to be related to shell formation by participating in calcium metabolism (data not shown). Considering the incompatibility between shell matrix protein genes with other functional categories and the possibility that many matrix proteins have not been identified yet (Zhang and Zhang 2006), there is a good chance that we would find possible biomineralization-related genes in the S category.
The gene ontology analysis and the annotation from the KEGG database provided further clues to the functions of the unigenes. The pathways revealed in the KEGG annotation indicated interactions between genes in C. farreri. The unigenes annotated to the calcium signaling pathway could play an important role in shell formation and therefore will be key subjects for future research.
To obtain more information about biomineralization in C. farreri, we searched for matrix proteins in the mantle transcriptome of C. farreri. Homologous unigenes of matrix proteins, such as MSP-1, Shematrin-2, Aspein, and Prisilkin-39, were found. Among them, the expressions of unigenes homologous with MSP-1 were especially high. Compared with the expression of other homologs, this high level of expression suggested that MSP-1 may play crucial roles in the shell formation of C. farreri, making them our primary focus in subsequent research. Remarkably, MSP-1 was only identified in the close relative of C. farreri, P. yessoensis before (Sarashina and Endo 1998). Although biomineralization researches were performed in numerous molluscs, no homologous proteins were identified in other species ever since MSP-1 was first reported. This discovery of MSP-1 in our study, its unusually high expression together with its exclusive presence in two scallops implied that MSP-1 may be expressed mainly in scallops and have essential roles in their shell formation. This further suggested that the shell formation mechanisms between scallops and other molluscs, like pearl oysters, are distinct, and the evolution of shell formation between species is relatively rapid.
Nevertheless, homologs of other typical calcite formation-related matrix protein genes, such as the MSI31 and Prismalin-14, were not identified in C. farreri. One possibility is that the expression level of these proteins in C. farreri mantle is too low to be detected. Another explanation is that there are no such genes in C. farreri, which further implies that the shell formation mechanism of C. farreri differs from other mollusks.
Unigenes homologous to other proteins participating in biomineralization, such as chitin synthase, carbonic anhydrase, G protein, and calcineurin B, were also found. Chitin synthase, a transmembrane glycosyltransferase that synthesizes chitin and an important organic component in the shell, has been proven to be a key element in biomineralization (Weiss et al. 2006). Carbonic anhydrase is another enzyme whose activity was revealed to be pivotal in shell formation by studying the role of carbonic anhydrase in shell regeneration (Stolkowski 1951). The discovery of these genes will help us to form a more complete picture of shell formation process in C. farreri.
In our alignments, we noticed that most of the matrix protein genes with homologies (10 out of 17) participated in the formation of calcite shell layers. Moreover, only three proteins related to aragonite formation were found in our study. Although it is possible that certain existing genes may not be detected in the alignment because of their low level of expression or poor homology to the query proteins, we believe that this result may indicate that in C. farreri, the proportion and variety of shell matrix proteins related to aragonite formation is limited compared with those related to calcite formation.
Previous studies confirmed that the adult shells of C. gigas and P. yessoensis were mainly composed of calcite, with only a small proportion of the shell comprising aragonite (Lee et al. 2006; Lin et al. 2010). In our previous study of the observation and analysis of the C. farreri shell using scanning electron microscopy and Raman spectrum (data not shown), we found that, similar to C. gigas and P. yessoensis, most of the C. farreri shell is composed of calcite, while only a small and thin layer inside of the shell near the adductor area is made of aragonite. This result is consistent with our discovery of few or no genes related to aragonite in the mantle transcriptome. Taking into consideration the shell composition and transcriptome alignment results, it is apparent that there is a positive correlation between the proportions of calcium carbonate polymorphs and the amount and variety of corresponding matrix proteins in C. farreri.
In previous biomineralization research, most attention focused on Pinctada fucata. P. fucata is a typical pearl oyster, which has a remarkable nacreous layer in addition to a prismatic layer, and the quantities of calcite and aragonite in its shell are similar (Sudo et al. 1997). Numerous matrix proteins were uncovered in P. fucata; both those related to calcite formation (such as Aspein, MSI31, MSI7, and Prismalin-14) and those related to aragonite formation (such as Nacrein, Pearlin, MSI60, and N14), and the amounts of both types of proteins were similar (Marin et al. 2008). This positive relationship between the proportions of calcite and aragonite in shells and the amount and variety of corresponding matrix proteins in P. fucata is consistent with the results of our research, thus supporting our deduction.
In the discovery of matrix proteins, their relationship to certain polymorphs of calcium carbonate (calcite or aragonite) was explored individually through various functional experiments. Some of them are considered to be related to the formation of calcite and some to aragonite (Marin et al. 2008). From another perspective, the positive correlation found in our research and in P. fucata supported the presumption that the formation of different kinds of calcium carbonate (calcite and aragonite) in molluskan shells is regulated by different groups of proteins. This may further imply that there are distinct models for the formation of these two polymorphs of calcium carbonate.
In our study, 17 unigenes were found to fulfill the criteria as encoding potential biomineralization-related proteins. These unigenes represent ideal materials for further study. To test the accuracy of this prediction and explore their expression distributions functions, further experiments, such as PCR amplification, RT-PCR assay, in situ hybridization, and RNAi, are required. These experiments have already been incorporated into our future research plan.
In sum, based on the sequencing and annotation of C. farreri mantle transcriptome, we identified 53 unigenes homologous with reported matrix proteins and 171 homologies with proteins involved in general processes of shell formation. In addition, we identified 17 unigenes potentially involved in biomineralization. The results indicate that the variety of matrix proteins is related to shell composition. Moreover, our research suggests that the formation of different polymorphs of calcium carbonate in molluskan shells may be regulated by different groups of proteins. The data also support the existence of diverse models in the formation of calcite and aragonite.
References
Belcher AM, Wu XH, Christensen RJ, Hansma PK, Stucky GD, Morse DE (1996) Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature 381:56–58
Conesa A, Gotz S et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Eisenhaber B, Bork P, Eisenhaber F (1998) Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng 11(12):1155–1161
Eisenhaber B, Bork P, Eisenhaber F (1999) Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292(3):741–758
Eisenhaber B, Bork P, Yuan Y et al (2000) Automated annotation of GPI anchor sites: case study C.elegans. Trends Biochem Sci 25(7):340–341
Emanuelsson O, Nielsen H, Brunak S et al (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Fang D, Xu G, Hu Y, Pan C et al (2011) Identification of genes directly involved in shell formation and their functions in pearl oyster, Pinctada fucata. PLoS One 6(7):e21860
Grabherr MG, Haas BJ et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Iseli C, Jongeneel C et al (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 138–148
Jackson D, McDougall C, Woodcroft B et al (2010) Parallel evolution of Nacre building gene sets in molluscs. Mol Biol Evol 27(3):591–608
Kong Y, Jing G, Yan Z, Li C, Gong N et al (2009) Cloning and characterization of Prisilkin-39, a novel matrix protein serving a dual role in the prismatic layer formation from the oyster Pinctada fucata. J Biol Chem 284:10841–10854
Lee S, Kim Y, Choi H et al (2006) Primary structure of myostracal prism soluble protein (MPSP) in oyster shell, Crassostrea gigas. Protein J 25:288–294
Lin A, Ding X, Xie Z et al (2010) Microstructure of Patinopecten yessoensis (scallop) shell and correlations with functions. J Chin Ceram Soc 38(3):504–509
Lowenstam H (1981) Minerals formed by organisms. Science 211:1126–1131
Lowenstam H, Weiner S (1989) On biomineralization. Oxford University Press, New York
Marin F, Luquet G, Marie B et al (2008) Molluscan shell proteins: primary structure, origin, and evolution. Curr Top Dev Biol 80:209–276
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31246
Miao J, Pan L, Liu N et al (2011) Molecular cloning of CYP4 and GSTpi homologues in the scallop Chlamys farreri and its expression in response to benzo[α]pyrene exposure. Mar Genomics 4:99–108
Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T et al (1996) A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci U S A 93:9657–9660
Mutz KO, Heilkenbrinker A, Lönne M et al (2012) Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol
Newman AM, Cooper JB (2007) XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinforma 8:382
Nielsen H, Engelbrecht J, Brunak S et al (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Petersen T, Brunak B et al (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Samata T, Hayashi N, Kono M, Hasegawa K, Horita C et al (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett 462:225–229
Sarashina I, Endo K (1998) Primary structure of a soluble matrix protein of scallop shell: implications for calcium carbonate biomineralization. Am Mineral 83:1510–1515
Sarashina I, Endo K (2001) The complete primary structure of molluscan shell protein 1 (MSP-1), an acidic glycoprotein in the shell matrix of the scallop Patinopecten yessoensis. Mar Biotechnol 3:362–369
Shi Y, Yu C, Gu Z et al (2013) Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes. Mar Biotechnol 15:175–187
Shumway S (1991) Scallops: biology, ecology and aquaculture. Amsterdam, Elsevier Science
Stolkowski J (1951) Essai sur le determinisme des forme s mineralogiques du calcaire chez les etres vivants (calcair es coquilliers). Ann Inst Oceanogr. N.S. 26, 1–113
Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nakashima K, Takahashi T (1997) Structures of mollusc shell framework proteins. Nature 387:563–564
Sunyaev SR, Eisenhaber F, Rodchenkov IV et al (1999) PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng 12(5):387–394
Suzuki M, Murayama E, Inoue H, Ozaki N, Tohse H et al (2004) Characterization of Prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem J 382:205–213
Taylor JD (1973) The structural evolution of the bivalve shell. Palaeontology 16(3):519–534
Tsukamoto D, Sarashina I, Endo K (2004) Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem Biophys Res Commun 320:1175–1180
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Weiss IM, Schnönitzer V, Eichner N et al (2006) The chitin synthase involved in marine bivalve mollusk shell formation contains a myosin domain. FEBS Lett 580:1846–1852
Werner GDA, Gemmell P, Grosser S et al (2013) Analysis of a deep transcriptome from the mantle tissue of Patella vulgata Linnaeus (Mollusca: Gastropoda: Patellidae) reveals candidate biomineralising genes. Mar Biotechnol 15(2):230–243
Xiao J, Ford SE, Yang H, Zhang G, Zhang F et al (2005) Studies on mass summer mortality of cultured zhikong scallops (Chlamys farreri Jones et Preston) in China. Aquaculture 250:602–615
Yano M, Nagai K, Morimoto K, Miyamoto H (2007) A novel nacre protein N19 in the pearl oyster Pinctada fucata. Biochem Biophys Res Commun 362:158–163
Ye J, Fang L et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Zhang C, Zhang R (2006) Matrix proteins in the outer shells of molluscs. Mar Biotechnol (NY) 8:572–586
Zhang Y, Xie L, Meng Q, Jiang T, Pu R et al (2003) A novel matrix protein participating in the nacre framework formation of pearl oyster, Pinctada fucata. Comp Biochem Physiol Part B: Biochem Mol Biol 135:565–573
Zhang C, Xie L, Huang J, Liu X, Zhang R (2006) A novel matrix protein family participating in the prismatic layer framework formation of pearl oyster, Pinctada fucata. Biochem Biophys Res Commun 344:735–740
Zhang X, Zhao C, Huang C, Duan H et al (2011) A BAC-based physical map of Zhikong scallop (Chlamys farreri Jones et Preston). PLoS One 6(11):e27612
Zhang G, Fang X, Guo X et al (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54
Zhao X, Wang Q, Jiao Y et al (2012) Identification of genes potentially related to biomineralization and immunity by transcriptome analysis of pearl sac in pearl oyster Pinctada martensii. Mar Biotechnol 14(6):730–739
Acknowledgments
This study was funded by the National High Technology Research and Development Program of China (863 Program, 2012AA092204).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1
(DOC 49 kb)
Rights and permissions
About this article
Cite this article
Shi, M., Lin, Y., Xu, G. et al. Characterization of the Zhikong Scallop (Chlamys farreri) Mantle Transcriptome and Identification of Biomineralization-Related Genes. Mar Biotechnol 15, 706–715 (2013). https://doi.org/10.1007/s10126-013-9517-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-013-9517-0