Abstract
In zebrafish and other vertebrates, primordial germ cells (PGCs) are a population of embryonic cells that give rise to sperm and eggs in adults. Any type of genetically manipulated lines have to be originated from the germ cells of the manipulated founders, thus it is of great importance to establish an effective technology for highly specific PGC-targeted gene manipulation in vertebrates. In the present study, we used the Cre/loxP recombinase system and Gal4/UAS transcription system for induction and regulation of mRFP (monomer red fluorescent protein) gene expression to achieve highly efficient PGC-targeted gene expression in zebrafish. First, we established two transgenic activator lines, Tg(kop:cre) and Tg(kop:KalTA4), to express the Cre recombinases and the Gal4 activator proteins in PGCs. Second, we generated two transgenic effector lines, Tg(kop:loxP-SV40-loxP-mRFP) and Tg(UAS:mRFP), which intrinsically showed transcriptional silence of mRFP. When Tg(kop:cre) females were crossed with Tg(kop:loxP-SV40-loxP-mRFP) males, the loxP flanked SV40 transcriptional stop sequence was 100 % removed from the germ cells of the transgenic hybrids. This led to massive production of PGC-specific mRFP transgenic line, Tg(kop:loxP-mRFP), from an mRFP silent transgenic line, Tg(kop:loxP-SV40-loxP-mRFP). When Tg(kop:KalTA4) females were crossed with Tg(UAS:mRFP) males, the hybrid embryos showed PGC specifically expressed mRFP from shield stage till 25 days post-fertilization (pf), indicating the high sensitivity, high efficiency, and long-lasting effect of the Gal4/UAS system. Real-time PCR analysis showed that the transcriptional amplification efficiency of the Gal4/UAS system in PGCs can be about 300 times higher than in 1-day-pf embryos. More importantly, when the UAS:mRFP-nos1 construct was directly injected into the Tg(kop:KalTA4) embryos, it was possible to specifically label the PGCs with high sensitivity, efficiency, and persistence. Therefore, we have established two targeted gene expression platforms in zebrafish PGCs, which allows us to further manipulate the PGCs of zebrafish at different levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In vertebrates, the primordial germ cells (PGCs) are the common ancestors of spermatozoa and oocytes. PGCs are distinct from somatic cells during early embryonic development, and they undergo dynamic migration, proliferation, and differentiation, resulting in final formation of mature oocyte and sperm in adults which could transmit genetic information between generations through reproduction. By using the zebrafish as an animal model, previous studies have shown that a batch of factors contribute to the early specification and development of PGCs. For instance, vasa was the first PGC-specific factor that was found in zebrafish (Braat et al. 2000; Yoon et al. 1997), nanos-related gene is required for the proper migration and survival of zebrafish PGCs (Koprunner et al. 2001), and dead end is necessary for zebrafish PGCs migration and survival through mainly counteracting miRNA mediated silencing (Ketting 2007; Kedde et al. 2007; Weidinger et al. 2003; Liu and Collodi 2010). Besides, the PGC-originated cells contribute to and are integrated in later gonad development (Slanchev et al. 2005; Youngren et al. 2005). PGCs are ideal target cells for genetic manipulation since they could be specifically labeled, removed and isolated during early development (Koprunner et al. 2001; Saito et al. 2008; Ciruna et al. 2002; Weidinger et al. 2003). Most importantly, they will give rise to adult gametes which provide genetic materials necessary to form a whole organism. However, as an excellent model for studies of vertebrate development and genetics, zebrafish still lacks a highly efficient and highly specific PGC-targeted gene expression platform under conditional control.
A group of DNA recombination induced activation systems, i.e., Cre/loxP, Flp/FRT and PhiC31 att/int systems, utilizes tissue-specific expressed recombinases to direct recombination between pairs of recognition DNA targets and to activate a gene-of-interest in specific tissues (Branda and Dymecki 2004). Among those, the Cre/loxP system is the one that was discovered the earliest and applied the most widely (Abremski et al. 1983; Shaikh and Sadowski 1997; Hoess and Abremski 1985). Cre/loxP system has been applied to genetic modification and expression induction in various animals and plants, including mice (Lakso et al. 1992; Gu et al. 1994), Drosophila (Siegal and Hartl 1996) and Arabidopsis (Vergunst et al. 1998). In 2005, functionality of the Cre/loxP system in zebrafish was demonstrated by injection of Cre mRNA in early embryos (Langenau et al. 2005; Pan et al. 2005). Since then, several Cre transgenic zebrafish lines have been established using heat shock promoter (Yoshikawa et al. 2008; Thummel et al. 2005; Le et al. 2007) and promoters specific for a few cell types or tissues, such as hematopoietic progenitors (Wang et al. 2008), neural progenitors (Seok et al. 2010; Kroehne et al. 2011), and oocytes (Liu et al. 2008). Those studies demonstrate that the Cre/loxP system works in zebrafish either after heat shock or in certain tissues. Our previous studies also demonstrated that a pair of mutated loxP sites could be used for site-directed gene integration in zebrafish (Liu et al. 2007). Nevertheless, there has been no application of the Cre/loxP system in zebrafish PGCs, an ideal cell type for genetic manipulation. By using monomer red fluorescent protein (mRFP) as a reporter and askopos (kop) promoter as an effective germ cell-targeted promoter (Blaser et al. 2005), we report our use of the Cre/loxP system for PGC-targeted manipulation of mRFP expression in zebrafish.
Gain-of-function approaches are commonly used for genetic analysis in animal models. Zebrafish is often manipulated at embryonic stage through injection the mRNA encoding proteins of interest. However, it is difficult to confine the mRNA expression in specific tissue and later developmental stages of zebrafish. Thus, mRNA overexpression method is inappropriate for gene function analysis in a tissue-specific and long lasting manner in zebrafish. Fortunately, the use of the Gal4/UAS system in zebrafish can overcome these two shortcomings, as tissue-specific expression of Gal4 transcriptional activator could restrict the expression of target gene in specific tissues, and lasting activation of UAS sequence by Gal4 protein could drive durable gene expression in target tissues. The Gal4/UAS system usually utilizes two transgenic animal lines, an activator line and an effector line. In the activator line, tissue-specific promoter/enhancer is used to drive expression of Gal4 transcriptional activator in given types of cells. Whereas, in the effector line, upstream activation sequence (UAS) is used to drive the gene of interest, and the UAS sequence could be merely recognized and activated by the Gal4 transcriptional activator. Generally, the gene of interest is transcriptionally silent in the effector line, and it will be tissue specifically activated in the transgenic offspring once the effector line is crossed with the activator line (Guarente et al. 1982; Traven et al. 2006; Carey et al. 1989; Giniger et al. 1985; Phelps and Brand 1998). The Gal4/UAS system was firstly utilized in the genetic studies of Drosophila and it has been popularly used in this animal model. Until recently, zebrafish has taken advantage of the Gal4/UAS system for promoter trapping and for targeted gene expression in a few tissues (Davison et al. 2007; Asakawa et al. 2008; Scott et al. 2007). In consideration of the activation efficiency and possible toxic effect of the heterogeneously expressed Gal4 protein, the Gal4/UAS system was optimized in zebrafish (Distel et al. 2009). In Distel et al.’s study, a fusion construct containing Kozak translational upstream sequence and a Gal4 protein with attenuated repeats of the VP16 transactivator (TA) core sequence, KalTA4, proved to be highly potent but low in toxicity. In the present study, we utilized the optimized Gal4/UAS system for targeted expression in zebrafish PGCs. We will also compare and discuss the application range of the Gal4/UAS and Cre/loxP systems in zebrafish PGCs.
Materials and Methods
Zebrafish
Embryos were obtained from the natural mating of zebrafish of the AB genetic background (from the China Zebrafish Resource Center, Wuhan, China) and maintained, raised, and staged as previously described (Kimmel et al. 1995; Westerfield 1995). The experiments involving zebrafish were performed under the approval of the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
Preparation of DNA Constructs
The PGC-specific Cre expression construct, pTol2(kop-Cre-UTRnos1, CMV-EGFP-SV40), is a Tol2 transposon-based, bipartite construct consisting of zebrafish askopos (kop) promoter and nanos1 (nos1) 3’ untranslated region (UTR)-regulated Cre expression cassettes as well as a cis-linked CMV promoter and SV40 poly(A)-regulated EGFP reporter. We amplified nos1 3’UTR with primers of nos1-3’UTR-F and nos1-3’UTR-R from the kop-EGFP-F-nos1-3’UTR vector (Blaser et al. 2005) with introducing three enzyme sites BsiWI, MluI, and BspEI at the 5’-end of nos1 3’ UTR, as well as three enzyme sites XhoI, ApaI, and BglII at the 3’-end for subsequent insertion of other fragments, and subcloned it into the pMD18-T vector (TaKaRa). On the base of this construct, firstly, we inserted the left arm of Tol2 amplified with primers of TL-F and TL-R from pDestTol2pA2 (Urasaki et al. 2006; Kwan et al. 2007) at the BsiWI and MluI sites. Secondly, we added kop promoter with one more enzyme site AscI at the 3’-end amplified with primers of kop-F and kop-R from the kop-EGFP-F-nos1-3’UTR vector (Blaser et al. 2005) at the MluI and BspEI sites. Thirdly, we fused Cre open reading frame (ORF) amplified with primers of Cre-F and Cre-R from pBS185 CMV-Cre vector (Addgene) at the AscI and BspEI sites. Then the reporter cassette (CMV:EGFP:SV40) amplified with primers of CES-F and CES-R from pCMVEGFP (Liu et al. 2005) was inserted at XhoI and ApaI sites. Finally, the right arm of Tol2 amplified with primers of TR-F and TR-R from pDestTol2pA2 (Urasaki et al. 2006) was added at the ApaI and BglII sites. The resulted DNA construct was herein named as pkop:Cre.
The loxP-containing construct, pTol2(kop-loxP-SV40-loxP-mRFP-UTRnos1, CMV-EGFP-SV40), was constructed by replacing the Cre ORF of pkop:Cre with two loxP sites flanked SV40 poly(A) sequence and one mRFP ORF. The loxP-flanked SV40 poly(A) fragment was obtained by PCR amplification using the two primers, loxpSV40-F and loxpSV40-R, containing synthetic loxP sequences, and was subcloned into the pMD18-T vector (TaKaRa), resulting in the pMD18-T-loxP-SV40-loxP construct. The loxP-flanked SV40 poly(A) fragment and mRFP ORF were fused into a strand of DNA with AscI and BspEI at the two ends using four primers, lsl-mRFP-F, lsl-mRFP-m-R, lsl-mRFP-m-F and lsl-mRFP-R, by overlap extension PCR (Horton et al. 1989). The PCR product was cut with AscI and BspEI, and was inserted into the pkop:Cre plasmid from which the Cre ORF was removed, resulting in the pkop:loxP-SV40-loxP-mRFP construct.
The PGC-specific Gal4 expression construct, pTol2(kop-KalTA4-UTRnos1, CMV-EGFP-SV40), was also constructed on the base of the pkop:Cre plasmid. We amplified KalTA4 ORF with primers of KalTA4-F and KalTA4-R from the construct TK5xC (Distel et al. 2009) with AscI and BspEI at the ends, and replaced the Cre ORF of pkop:Cre with KalTA4 ORF at the AscI and BspEI sites, resulting in the pkop:KalTA4 construct. The UAS effector construct, pTol2(UAS-mRFP-UTRnos1, CMV-EGFP-SV40), was constructed using a similar construction as the pkop:loxP-SV40-loxP-mRFP construct. The 4× UAS sequences and mRFP ORF were fused into a strand of DNA with the addition of MluI and BspEI at the ends using four primers, UAS-mRFP-F, UAS-mRFP-m-R, UAS-mRFP-m-F and 4UAS-mRFP-R, by overlap extension PCR, and the kop-Cre-nos1 expression cassette of pkop:Cre was replaced by the UAS-mRFP-nos1 fusion fragment at the MluI and BspEI sites, resulting in the pUAS:mRFP construct.
For transient expression assay, the construct pUAS:mRFP-SV40 was constructed by replacing the CMV promoter of PCS2+ vector (Addgene) with UAS–mRFP fusion fragment amplified with primers of UASmRFP-F and UASmRFP-R from pUAS:mRFP construct at the SalI and XhoI sites. The construct pCMV:loxP-SV40-loxP-mRFP-SV40 was constructed by amplifying loxp-SV40-loxp-mRFP fusion fragment with adding the BamHI and XhoI sites at the ends with primers of loxpmRFP-F and loxpmRFP-R from pkop:loxP-SV40-loxP-mRFP, and the PCR product was inserted into PCS2+ vector (Addgene) at the BamHI and XhoI sites.
Microinjection and Generation of Transgenic Zebrafish Lines
To generate transgenic lines, zebrafish embryos were injected with approximately 1 nL of a DNA/RNA solution containing 25 ng/μL Tol2 transposase mRNA and 25 ng/μL Tol2-based transgenic construct at one-cell stage (Urasaki et al. 2006). The embryos showing EGFP expression were screened and raised to adulthood and the 3-month-old fish (F0) were crossed to wildtype fish for analyzing germline transmission by screening the offspring under a fluorescence microscope (Olympus MVX10). F1 embryos expressing EGFP were raised to adulthood and were then screened in the same way to establish stable transgenic lines. For transient expression assay, DNA construct (25 ng/μL) was directly injected into one-cell stage embryos, and the injected embryos were allowed to develop at 28 °C for analysis of fluorescent protein expression.
PCR Analysis and DNA Sequencing
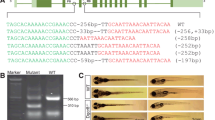
To verify Cre-mediated recombination events in Tg(kop:loxP-SV40-loxP-mRFP) after crossing with female Tg(kop:Cre), PCR was performed with primers of kop-mRFP-F1 (F1) from zebrafish kop promoter and kop-mRFP-R1 (R1) from the mRFP ORF. To test the efficiency of the recombination in different tissues of the hybrid founder fish, a pair of primers were designed and used for PCR analysis, primer kop-SV40-F2 (F2) locating in zebrafish kop promoter and primer kop-SV40-R2 (R2) locating in the loxp-flanked SV40 poly(A) sequence. In addition, to confirm the presence of Cre transgene fragment, genomic DNA from different tissues was examined by PCR with Cre-specific primers, Cre-F3 and Cre-R3. The following cycling conditions were used: denaturation of the DNA template at 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 1 min, and a final 72 °C extension for 10 min. All the PCR products were analyzed by agarose gel electrophoresis. The PCR products resulting from primers F1 and R2 were gel-extracted, subcloned into the pMD18-T vector (TaKaRa), and sequenced.
RNA Whole-Mount In Situ Hybridization
To generate Kal4TA4, Cre, and mRFP antisense RNA probes for whole-mount in situ hybridization, we used three forward primers: Kal4TA4-specific primer (Probe-KalTA4-F), Cre-specific primer (Probe-Cre-F), mRFP-specific primer (Probe-mRFP-F), and the same reverse primer of T3-nos1 3’UTR-R which introduced T3 promoter sequences in the 5’-end from nos1 3’UTR sequences, for PCR amplification of partial sequences of Kal4TA4, Cre, and mRFP from pkop:KalTA4, pkop:Cre, and pkop:loxP-SV40-loxP-mRFP, respectively. The PCR products served as templates to synthesize antisense RNA probes using the digoxigenin RNA labeling kit (Roche) and T3 RNA polymerase (Promega), and purified using mini-Spin Columns (Sigma). One-color whole-mount in situ hybridization was carried out as previously described (Thisse et al. 2004) and NBT/BCIP substrate solution (Roche) was used in the color reaction to detect the probes.
Quantitative Real-Time PCR
Fifty embryos at 1 day post-fertilization (pf) from testing lines were collected and homogenized in TRIZOL reagent (Invitrogen) to isolate total RNA. For the quantitative RT-PCR analysis, 1 μg of total RNA was used as templates for the first strand cDNA synthesis with oligo(dT) and Superscript II (Invitrogen). Then quantitative real-time PCR was carried out with SYBR green dye and TaqMan analyses were performed using Applied Biosystems 7900 Real-Time PCR System according to the manufacturer’s directions. Relative expression levels of EGFP or mRFP were calculated using 2^(-delta delta C(T)) method with β-actin as the internal control. Variance analysis (one-way ANOVA) was employed for comparing the mean differences between the experimental groups. A P value < 0.05 was considered statistically significant (Livak and Schmittgen 2001). The primer sequences used to perform the PCR amplication are listed in Table S1.
Results
Generation of Transgenic Lines for Cre/loxP System
In order to test the Cre/loxP system in zebrafish PGCs, we first designed and constructed a PGC-specific Cre expression vector, pkop:Cre (Fig. 1a). The construct contains two bicistronic expression cassettes, kop:Cre:nos1 in which the Cre recombinase gene is driven by germ cell-specific kop promoter (Blaser et al. 2005) and nanos1 3’UTR (Koprunner et al. 2001), and CMV:EGFP:SV40 in which enhanced green fluorescent protein (EGFP) gene is driven by CMV promoter and SV40 poly(A) signal. The CMV:EGFP:SV40 expression cassette was used as a fluorescent reporter to facilitate transgenic screening. Among 40 randomly selected transgenic founders, four produced EGFP-positive F1 embryos (Table 1). We further raised one batch of transgenic F1 embryos to establish a homozygous transgenic line, Tg(kop:Cre) ihb7.
Generation of transgenic zebrafish lines of the Cre/loxP system. Schema of the kop:Cre construct (a). Schema of the kop:loxP-SV40-loxP-mRFP construct (b). Spatio-temporal expression pattern of cre mRNA in four-cell stage (c), shield stage (d), early somite stage (e), and 1 day pf (f) embryos of the Tg(kop:Cre) line. Arrows in c–f point to the PGC specifically expressed cre mRNA signals
To assay the expression of Cre mRNA in Tg(kop:Cre) embryos, we fixed the transgenic embryos at different stages and performed whole mount in situ hybridization using cre antisense probe. As shown in Fig. 1c, prominent signals were visible in the germplasm at the cleavage planes in the four-cell stage embryos. Later in development, cre-expressed cells distributed in four separate groups near the margin of shield stage embryos (Fig. 1d), migrated to the dorsal side to form two groups on both sides of the notochord during somitogenesis (Fig. 1e), and located in future genital ridge of 1-day-pf embryos (Fig. 1f). These revealed that the Tg(kop:Cre) embryos could specifically express Cre recombinase in the PGCs from early stage during development.
We then designed and constructed a loxP transgene construct, pkop:loxP-SV40-loxP-mRFP, which also contains two bicistronic expression cassettes (Fig. 1b). The first expression cassette contains a loxP flanked SV40 poly(A) signal between the kop promoter and the mRFP gene, while the second one is a CMV:EGFP:SV40 fluorescent reporter. Among 48 transgenic founders, we identified three that produced green fluorescent F1 embryos (Table 1). A homozygous transgenic line, Tg(kop:loxP-SV40-loxP:mRFP) ihb10, which carries the above construct was established. The transgenic line showed strong EGFP expression in the whole body but no mRFP expression in any tissue (data not shown).
High-Efficient PGC-Targeted Excision of SV40 Stop Sequence in Cre/loxP Hybrid Fish
To determine whether Cre mediated loxP recombination in the germ cells of zebrafish, we conducted cross-hybridization between homozygous Tg(kop:Cre) females and homozygous Tg(kop:loxP-SV40-loxP-mRFP) males for the maternal effect of the kop promoter (Blaser et al. 2005). The hybrid embryos were examined for EGFP and mRFP expression. Although all the embryos were shown to be EGFP positive in the whole body, no red fluorescence could be observed in those embryos, which should be due to the strict maternal activity of the kop promoter that drives mRFP expression (Blaser et al. 2005). These hybrid embryos were raised to adulthood and they were designated as the hybrid founders. The genomic DNA from various tissues of the hybrid founders was prepared, and PCR was carried out to examine the excision of SV40 stop sequences between two loxP sites with primer pair F1 and R1, which locate in the kop promoter and mRFP coding region, respectively. The expected amplification bands before and after Cre-mediated recombination are 841 bp and 599 bp, respectively (Fig. 2a). In all of the somatic tissues examined, both the 841-bp pre-excision fragment and the 599-bp post-excision fragment could be detected, suggesting Cre-mediated recombination in the hybrid founders in various cell types (Fig. 2b). In the sperm or oocyte samples, intriguingly, only the post-excision fragment of 599 bp could be detected. Sequencing of the 841-bp and 599-bp fragments revealed that the loxP-flanked SV40 transcriptional stop sequences were precisely excised from the transgenic loci in the germ cells (Fig. 2c). To confirm the excision efficiency of the Cre-mediated recombination in the hybrid founders, additional PCR was performed with primer pair F2 and R2, which locate in the kop promoter and SV40 poly(A) sequence, respectively (Fig. 2a). As expected, a 488-bp unexcised SV40 poly(A) fragment existed in all types of somatic tissues, whereas no specific amplicon could be obtained from the genomic DNA of the sperm or oocyte (Fig. 2b). This confirmed that the loxP-flanked SV40 transcriptional stop sequence was completely removed in the germ cells, and thus Cre-mediated recombination achieved at an efficiency of 100 % in the germline of the hybrid founders. Therefore, the hybrid founders reproduced from Tg(kop:Cre) females and Tg(kop:loxP-SV40-loxP-mRFP) males could be considered as a special transgenic fish, which contains Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP) somatic cells and Tg(kop:cre, kop:loxP-mRFP) germ cells (Fig. 3a). In contrast, Cre-mediated recombination was not detected in the hybrid founders resulted from the cross between male Tg(kop:Cre) fish and female Tg(kop:loxP-SV40-loxP-mRFP) fish (data not shown).
Cre recombinase mediates efficient excision of SV40 stop signal in zebrafish germline. a Schema of the polymerase chain reaction primer positions and product size. b Cre-mediated recombination in transgenic fish was detected by PCR analysis. PCR was performed for different tissues of the hybrid founders, Tg(kop:cre, kop:loxP-SV40-loxP-mRFP), using primers F1 and R1 with expected sizes of PCR products before and after Cre-mediated excision. Primers F2 and R2 amplified 488-bp unexcised SV40 poly(A) fragment. Presence of Cre DNA was also amplified by PCR using a pair of Cre-specific primers. The 599-bp recombined fragment and 841-bp full-length transgene fragment of kop:loxP-SV40-loxP-mRFP existed in all types of somatic cells in Tg(kop:cre, kop:loxP-SV40-loxP-mRFP). However, in the oocyte and sperm samples, only the 599-bp recombined fragment can be detected but not 841-bp full-length transgene fragment and 488-bp unexcised SV40 poly(A) fragment. PCR was also performed to check the presence of the kop:Cre transgene fragment, using Cre-specific primers, Cre-F3 and Cre-R3. Samples: DL2000 DNA marker (Fermentas); tail fin; muscle; eye; gills; heart; liver; air bladder; intestines; brain; spleen; skin; oocyte; sperm; positive (PCR control with pkop:loxP-SV40-loxP-mRFP or pkop:Cre plasmids); negative (wildtype embryos). c Sequence traces from genotyping. After excision by Cre recombinase, the stop signal and one loxP site was precisely excised from the genomic DNA in the hybrid embryos
Conditional activation of PGC-specific mRFP expression in the transgenic zebrafish by the Cre/loxP system. a The schema for the genotype of the Cre/loxP system. The genotype is Tg(kop:Cre, kop:loxp -mRFP) in the germ cells of progeny from a cross between female Tg(kop:Cre) and Tg(kop:loxP-SV40-loxP-mRFP:nos1) males. b–e mRFP expression in the hybrid founders and the hybrid offspring resulted from the Cre/loxP system. Neither PGC-specific mRFP protein (b, b’) nor mRFP mRNA (c) was detected in the hybrid founder at 1 day pf. Weak PGC-specific expression of mRFP proteins (d, d’) and mRFP mRNA (e) was observed in the hybrid offspring at 1 day pf. Arrows point to the PGC specially expressed mRFP proteins (d) or mRNA signals (e)
To verify the germline-specific excision of loxP-flanked SV40 transcriptional stop sequence in Tg(kop:loxP-SV40-loxP-mRFP) offspring, the mRFP silent hybrid founders were incrossed, and the resulting embryos were checked for expression of mRFP in germ cells (Fig. 3a). Interestingly, although the hybrid founders did not show PGC-specific expression of mRFP by fluorescent observation (Fig. 3b, b’) and RNA in situ hybridization (Fig. 3c), the hybrid offspring showed PGC-specific expression of mRFP at both protein and mRNA levels (Fig. 3d, d’, e). Therefore, through a germline targeted Cre/loxP system, we were able to achieve massive production of transgenic embryos, Tg(kop:loxP-mRFP), from a transgenic silent line, Tg(kop:loxP-SV40-loxP-mRFP).
Generation of Transgenic Lines for Gal4/UAS System
In order to realize the PGC-targeted expression in zebrafish embryos by Gal4/UAS system, we first designed and constructed a PGC-specific Gal4 expression vector, pkop:KalTA4 (Fig. 4a). The construct contains two bicistronic expression cassettes, kop:KalTA4-nos1, in which the KalTA4 activator protein gene is driven by germ cell-targeted kop promoter and nanos1 3’UTR, and the fluorescent reporter CMV:EGFP-SV40. Among 45 randomly selected transgenic founders, four produced EGFP-positive transgenic F1 embryos (Table 1). We further raised one batch of transgenic F1 embryos and set up a homozygous line of Tg(kop:KalTA4) ihb8 . The expression of KalTA4 mRNA in Tg(kop:KalTA4) embryos was exactly similar to the expression of Cre mRNA in Tg(kop:cre) embryos as shown in Fig. 1c, d (data not shown).
PGC-specific mRFP expression in the embryos induced by the Gal4/UAS system. a Schema of the kop:KalTA4 construct. b Schema of the UAS:mRFP construct. c–f Weak expression of EGFP was observed in progeny of Tg(kop:EGFP) females from early somite stage (c), at 1 day pf (d), 3 days pf (e), but not at 9-day-pf embryos (f). g–n mRFP expression in the embryos from a cross between Tg(kop:KalTA4) female and Tg(UAS:mRFP) male. Strong expression of mRFP was detectable under a fluorescence microscope, from shield stage in PGCs (g, g’). Strong mRFP expression could be visualized at 1 day pf (h, h’), 3 days pf (i, i’), 9 days pf (j, j’). At 18 days pf, mRFP expression continued strongly in most gonad cells of some larvae (k, k’), and in a few gonad cells of other larvae (l, l’). mRFP expression was still detectable by 25 days pf in a few larvae (m, m’), and was undetectable by 30 days pf in all of the embryos (n, n’). o Comparison of mRFP and EGFP mRNA expression levels at 1 day pf among Tg(kop:EGFP) embryos, Tg(kop:Cre,kop:loxp-SV40-loxp-mRFP) embryos, and a cross between Tg(kop:KalTA4) female and Tg(UAS:mRFP) male by real-time PCR, using β-actin mRNA as internal control. The mRFP mRNA expression levels of embryos from incross of Tg(kop:Cre,kop:loxp-SV40-loxp-mRFP) and Tg(kop:EGFP) were similar (P > 0.05) to each other. Significant difference (P < 0.001) of mRFP mRNA expression levels was found between embryos from a cross between female Tg(kop:KalTA4) and male Tg(UAS:mRFP) with others. Arrows point to PGC specifically expressed EGFP or mRFP signals
We then designed and constructed a UAS-dependent transgene construct, pUAS:mRFP, which also contains two bicistronic expression cassettes (Fig. 4b). The first expression cassette UAS:mRFP-nos1 contains a four Gal4-binding-site (4 × UAS)-driven mRFP and nanos1 3’UTR, while the second one is a CMV:EGFP-SV40 fluorescent reporter. Among 36 transgenic founders, we screen out four which could produce EGFP-positive F1 embryos (Table 1). A homozygous transgenic line, Tg(UAS:mRFP) ihb9, that carries the above construct, was established. The transgenic line showed strong EGFP expression but no mRFP expression (data not shown).
High-Efficient PGC-Targeted Expression in Hybrid Embryos of Tg(UAS-mRFP) and Tg(kop:KalTA4)
Due to the maternal effect of the kop promoter (Blaser et al. 2005), the hybrid embryos resulted from Tg(kop:Cre) females, and Tg(kop:loxP-SV40-loxP-mRFP) males did not show PGC-specific mRFP expressions, although the loxP-flanked SV40 stop signal was removed from the transgene in the PGCs with high efficiency. Thus, we were curious about the hybrid embryos which resulted from Tg(kop:KalT4) and Tg(UAS:mRFP). Prior to testing this, we first generated several Tg(kop:EGFP) ihb8 lines that carried the kop:EGFP-nos1 expression construct (a gift of Prof. Raz) (Blaser et al. 2005). In all of the Tg(kop:EGFP) lines we tested, the PGC-specific EGFP expression was only detectable from early somite stage and lasted to 3 days pf under the fluorescent microscope in our lab (Fig. 4c–f). Given the activation potential of the Gal4/UAS system to amplify the expression of target genes downstream which has been used in many organisms, we were interested whether the Gal4/UAS system could drive and even amplify mRFP expression in zebrafish PGCs. In the embryos from a cross between Tg(kop:KalTA4) females and Tg(UAS:mRFP) males, intriguingly, PGC-specific mRFP expression was detectable under the same fluorescent microscope at as early as shield stage (Fig. 4g, g’), and the expression tended to be stronger and stronger along development (Fig. 4h–j, h’–j’). Strong mRFP expression could be visualized specifically in PGCs of the hybrid embryos from 1 day pf and lasted till 15 days pf. Later, the intensity of mRFP varied among different larvae from a few fluorescent cells in the gonad to almost whole fluorescing gonad at 18 days pf. mRFP was still detectable by 25 days pf in most embryos, and is undetectable by 30 days pf (Fig. 4m, m’, n, n’).
In order to compare the transcriptional levels among different activation systems, i.e., the Cre/loxP and Gal4/UAS systems, the mRFP transcriptional levels at 1 day pf among embryos from Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP) female and a cross between female Tg(kop:KalTA4) and male Tg(UAS:mRFP) were compared with the embryos of Tg(kop:EGFP) female by real-time PCR. When β-actin was used as an internal control, as expected, mRFP mRNA expression levels of the embryos from incross of Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP) was similar to the embryos of Tg(kop:EGFP) (P > 0.05), since both types of embryo were driven by the kop promoter and the nanos1 3’UTR. Interestingly, we found that the mRFP RNA transactivation level in the embryos from a cross between Tg(kop:KalTA4) females and Tg(UAS:mRFP) males were about 300-fold stronger than that in the Tg(kop:EGFP) embryos or the embryos from incross of Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP) (P < 0.001) (Fig. 4o). These demonstrate that the Gal4/UAS system could not only activate the gene expression but also dramatically amplify the expression level in zebrafish PGCs.
mRFP mRNA Spatio-temporal Expression Pattern in Cre/loxP and Gal4/UAS Hybrid Fish
The detection of mRFP in PGCs under a fluorescence microscope is not indicative of how early the PGC-specific Gal4 protein can drive transcription of mRFP under UAS, since accumulation of sufficient mRFP protein for visual detection is delayed with respect to initial mRFP transcription. Besides, it is possible that sites of mRFP expression are missed because mRFP expression level is undetectable by the eyes under a fluorescence microscope. To determine more accurately the spatio-temporal distribution of mRFP in the Cre/loxP- and Gal4/UAS-activated embryos, we observed the mRFP transcripts by whole-mount in situ hybridization on embryos from incross of Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP), and a cross between female Tg(kop:KalTA4) and male Tg(UAS:mRFP), and utilized the vasa expression as a control to mark the location of germplasm and PGCs (Fig. 5a–e). In the embryos from the incross of Tg(kop:Cre, kop:loxP-SV40-loxP-mRFP), the mRFP transcripts were provided maternally and enriched in the distal end of the first two cleavage furrows of the four-cell stage embryos (Fig. 5f), indicating that the kop promoter was already activated during oogenesis of the hybrid founders and the loxP flanked SV40 stop sequences were completely excised in the germline genome of the hybrid founders. The PGC-specific expression of mRFP was obvious in the following developmental stages (Fig. 5g–j). Whereas, in the embryos from a cross between Tg(kop:KalTA4) females and Tg(UAS:mRFP) males, the germplasm-specific mRFP mRNA was undetectable at the four-cell stage (Fig. 5k), and PGC-specific mRFP expression was only detectable starting at the sphere stage in four separate groups in the hybrid embryos (Fig. 5l). The mRFP-positive cells in all the testing embryos migrated and formed to four separate clusters, spaced around the embryo, and generally near the margin at shield stage (Fig. 5h, m), and then clustered into two groups on either side of notochord at somite stage (Fig. 5i, n). At 1 day pf, the mRFP transcripts located bilaterally where the yolk ball meets the yolk tube (Fig. 5j, o). The expression pattern of mRFP transcripts in both Cre/loxP and Gal4/UAS hybrid fish resembles that described for the zebrafish vasa mRNA, which is a specific marker for germ cells. These strongly supported that mRFP was expressed in PGCs of the hybrid embryos resulted from the two systems.
Spatio-temporal expression pattern of mRFP mRNA in the embryos resulted from Cre/loxP and Gal4/UAS systems. a Whole-mount in situ analysis was performed to determine the distribution of vasa (a–e) mRNA and mRFP mRNA in embryos from incross of Tg(kop:Cre,kop:loxp-SV40-loxp-mRFP)(F-J) and a cross between Tg(kop:KalTA4) female and Tg(UAS:mRFP) male (k–o) from four-cell stage to 1 day pf stage. The developmental stages are: four-cell stage (a, f, k), sphere stage (b, g, l), shield stage (c, h, m), early somite stage (d, i, n), and 1 day pf (e, j, o), respectively. The arrows point to the transcripts enriched to the cleavage furrows and the PGCs, respectively
Transient PGC-Targeted Expression of mRFP by Direct Injection of Different Expression Constructs
Since both the Cre/loxP and Gal4/UAS systems could specifically activate targeted gene expression in PGCs of the hybrid embryos, we then asked whether it is possible to activate mRFP expression in the activator embryos, i.e., Tg(kop:KalTA4) embryos or Tg(kop:Cre) embryos, by direct injection of effector constructs, such as UAS:mRFP-nos1 and UAS:mRFP-SV40, or CMV:loxP-SV40-loxP-mRFP-SV40. As expected, by injecting kop:EGFP-nos1 construct directly into wildtype embryos, it was impossible to observe PGC-specific expression of EGFP due to the maternal effect of the kop promoter (Fig. 6a). However, it was very efficient to label the PGCs of the host embryos with mRFP expression by injecting UAS:mRFP-nos1 or UAS:mRFP-SV40 constructs into Tg(kop:KalTA4) embryos, with the former one of PGC-specific expression of mRFP (Fig. 6b, red arrowhead) and the latter one of some ectopic expression of mRFP other than PGC-specific expression (Fig. 6c, white and red arrowhead). These indicate that the KalTA4 activator proteins were maternally expressed and thus could possibly activate the mRFP expression in somatic cells when UAS:mRFP-SV40 construct was injected. When CMV:loxP-SV40-loxP-mRFP-SV40 was injected into Tg(kop:Cre) embryos, PGC-specific expression of mRFP could be detected in all of the injected embryos, but most embryos showed ectopic expression of mRFP in somatic cells other than PGCs (Fig. 6d, white and red arrowhead). This result was consistent to the fact that cre mRNA was expressed maternally in the Tg(kop:Cre) embryos and thus it was only restricted to the PGCs during later developmental stages (Fig. 1c–f). The PGC-specific expression of mRFP in the Tg(kop:KalTA4) embryos injected with UAS:mRFP-nos1 was then analyzed in detail. The PGC-specific red fluorescence was visualized strongly and exclusively in PGCs from early somite stage in the injected embryos (Fig. 6e). With the embryonic development, mRFP was still specifically and strongly expressed in the PGCs of the injected embryos from 1 day pf to 9 days pf (Fig. 6f–h), indicating that by using the Gal4/UAS system, it is possible to visualize the zebrafish PGCs with high sensitivity and persistence by direct injection of a certain DNA construct.
Transient expression of fluorescent proteins in zebrafish embryos by injection of different expression constructs. a Schema of construct UAS:mRFP-SV40. b Schema of construct CMV:loxP-SV40-loxP-mRFP-SV40. c EGFP could not be detectable in wildtype embryos by injecting kop:EGFP construct. d, g–j mRFP was visualized strongly and specifically in the PGCs of Tg(kop:KalTA4) embryos after direct injection of pUAS:mRFP vector from early somite stage (g) to 9 days pf (j). e mRFP was not only detected in the PGCs but also in other somatic cells after injection of pUAS:mRFP-SV40 vector into Tg(kop:KalTA4) embryos. f mRFP was observed in PGCs and other types of cells in Tg(kop:Cre) embryos after injection of pCMV:loxp-SV40-loxp-mRFP:SV40 construct. The red arrows mark the PGC-specific mRFP expression, and the white arrows show the ectopic expression of mRFP in somatic cells
Discussion
Germ cells are unique and important for sexually reproducing eukaryotes, since they are the only cell population which could inherit genetic materials between generations. To study the development of germ cells has long been one of the focuses of cell and developmental biologists. Besides this, any type of genetically manipulated lines originated from the germ cells of the manipulated founders, thus it is of great importance to establish an effective technology for highly specific PGC-targeted gene manipulation in vertebrates. Cell type-specific genetic modification using the induction system is an invaluable tool for studies of distinct cell lineages. In the present study, we utilized two induction systems, the Cre/loxP recombination system and Gal4/UAS transcription system, and mRFP gene as a reporter, to realize high-efficient PGC-targeted gene expression, in the notable animal model, zebrafish.
In vertebrates, only a few germ cell-specific Cre transgenic lines have been established in the mouse model, which allow germ cell-specific genetic modifications to harbor visual markers to detect germ cells in vivo and to conduct conditional knockout in germ cells (Gallardo et al. 2007; Hammond and Matin 2009; Lomeli et al. 2000; Ohinata et al. 2005; Suzuki et al. 2008). However, the recombinant efficiencies induced by germ cell-expressed Cre in mice were varied between 25 % (for Nanos3-Cre) and 60 % (for TNAP-Cre). In zebrafish, although Cre-mediated global excision has been demonstrated by injection of cre mRNA into the zygotes of some transgenic lines, such as Tg(mylz2:loxP-EGFPloxP) gz3 (Pan et al. 2005) and Tg(rag2-loxP-dsRED2-loxP-EGFP-mMyc) (Langenau et al. 2005), there is no report of PGC-specific Cre lines and PGC-specific Cre-mediated recombination. The only germ cell type Cre transgenic zebrafish line is Tg(zp3:cre,krt8:rfp), which was constructed using oocyte-specific zp3 promoter and not suitable for studying the development of zebrafish PGCs and conducting PGCs manipulation (Liu et al. 2008). In the present study, we utilized an upstream kop promoter and a downstream 3’UTR of the nanos1 gene, which confer specific EGFP expression in PGCs in the Tg(kop:EGFP) transgenic embryos (Blaser et al. 2005), to drive PGC-specific expression of Cre in Tg(kop:cre). As a result, we observed the exquisite specificity of the cre mRNA expression pattern in the PGCs of the transgenic embryos. This PGC-specific Cre transgenic line of zebrafish will provide a platform to perform PGC-conditional gene function analysis and PGC-targeted gene modifications. In addition, to verify the function of PGC-specifically expressed Cre recombinase in Tg(kop:cre), we established a loxP transgenic line, Tg(kop:loxP-SV40-loxP-mRFP). By crossing the Tg(kop:cre) females and Tg(kop:loxP-SV40-loxP-mRFP) males, we demonstrated that the PGCs specifically expressed Cre provided by Tg(kop:cre) is capable of conditional activation of loxP-blocked genes in the hybrid embryos. In our study, the efficiency of Cre-mediated germline recombination achieved 100 %, which was strongly supported by two types of PCR analysis. In all of the previously described PGC-specific Cre transgenic mouse lines, the Cre-mediated recombination happened in other tissues besides the germ cells (Suzuki et al. 2008; Gallardo et al. 2007; Lomeli et al. 2000; Ohinata et al. 2005). In our study, Cre-meditated recombination was also detected in other somatic tissues, which was due to the maternal expression of Cre mRNA, just similar to the study of Tg(zp3:cre,krt8:rfp) (Liu et al. 2008). Nevertheless, we have generated a PGC-specific Cre transgenic line in zebrafish and demonstrated a PGC-specific recombination with an effeciency of 100 %. In future studies, it is possible to use a drug-inducible CreERT2 (John et al. 2008) to eliminate the maternal effect of Cre recombinase in Tg(kop:Cre).
Although many stable transgenic lines were established that express Gal4 in a tissue-specific manner with the development of an enhancer trapping system, which enabled investigators to regulate gene expression in specific cell types and tissues (Davison et al. 2007), no PGC-specific Gal4 transgenic line has been reported in zebrafish. In Drosophila, using the promoter and 3’ UTR of the nanos RNA, the nos–Gal4–VP16 transgene was generated for the activation of a germline-specific expression of targets (Van Doren et al. 1998). In the present study, we generated the Tg(kop:KalTA4) transgenic line, in which the KalTA4 activator was driven by the kop promoter and nanos1 3’UTR, and KalTA4 mRNA was expressed specifically in PGCs. To date, researchers have generated transgenic lines to label the PGCs in live embryos of zebrafish (Blaser et al. 2005; Fan et al. 2008; Knaut et al. 2002; Krovel and Olsen 2002). Nevertheless, these transgenic constructs and/or approaches still did not provide ideal tools for labeling, isolation, and genetic manipulation of zebrafish PGCs, because (1) direct injection of these constructs into zygotes could not distinctly label the PGCs, (2) the germ cell targeted promoter used in those studies is rather large in size and it is not convenient to use them for genetic manipulations, (3) the expression levels of fluorescent proteins driven by these constructs were not elevated enough for genetic manipulation (from our own experiments). In the present study, by utilizing the transcriptional amplification speciality of the Gal4/UAS system, we demonstrated the hybrid embryos resulted from Tg(kop:KalT4) females and Tg(UAS:mRFP) males showed germ cells specifically expressed mRFP from shield stage till 25 days pf. Interestingly, the level of PGC-specific mRFP expression in 1-day pf embryos is as high as 300 times in the Gal4/UAS hybrid embryos, so that strong mRFP expression could be visualized specifically in PGCs of the hybrid embryos resulted from the Gal4/UAS system. Although the expression delay of transgene induced by the Gal4/UAS system in zebrafish was demonstrated in a previous study (Zhan and Gong 2010), here we observed that mRFP mRNA could be detected in PGCs at sphere stage, close to the time when PGCs segregated (2.75 h pf), which suggests that it did not required a long recovery time for transcriptional activation when the Gal4/UAS system was used in zebrafish PGCs. Moreover, compared with that, the PGC-specific mRFP expression cannot be detectable at 9 days pf in the embryos of transgenic lines Tg(kop:EGFP), the hybrid embryos showed germ cell-specific mRFP expression from shield stage till 25 days pf, indicating the high sensitivity, high efficiency, and long-lasting effect of this system. More importantly, by direct injection of the UAS:mRFP-nos1 construct into the Tg(kop:KalTA4) zygotes, it was possible to label the PGCs of host embryos with mRFP from early somite stage till 9 days pf. To our knowledge, it is the first time that zebrafish PGCs could be exclusively and distinctly labeled by direct injection of an expression construct. Moreover, the small size of the UAS regulatory region (<100 bp) allows the expression cassette to be conveniently used as a genetic marker or a selection tool for further PGC manipulations.
In conclusion, we have established two induction systems, Cre/loxP recombination system and Gal4/UAS transcription system, to realize high-efficient PGC-targeted gene expression in zebrafish. These transgenic zebrafish lines will provide powerful tools to conduct conditional gene activation and inactivation systems for studies of gene function in PGCs. In addition, they have some other obvious applications. First, Tg(kop:Cre) would be useful to develop some conditional transgenic lines that may display early defects or mortality because of the global overexpression of certain functional genes. Second, as the Cre-mediated excision achieved an efficiency of 100 % in germ cells of the Cre/loxP hybrid founders, Tg(kop:Cre) can serve as a transgenic fish model to investigate the feasibility of self-excision of transgenes from transgenic fish to address the proprietary issue as well as ecological concerns on transgene contamination. As demonstrated in transgenic plants (Luo et al. 2007), it is possible to engineer transgenic fish to retain a beneficial phenotype from the transgene while the transgene is programmed to be excised from the germline by the germline-specific Cre/loxP system; thus the released transgenic fish will not be able to produce transgenic offspring even if they accidently escaped to a natural body of water. Third, owing to the strong and specific mRFP expression in PGCs of the hybrid embryos which resulted from Tg(kop:KalT4) and Tg(UAS:mRFP), it offers a useful tool for tracking and manipulating germ cells during zebrafish development. Finally, the expression cassette, UAS:mRFP-nos1, would be useful to facilitate screening of the candidates for germ line-transmitting founders if Tg(kop:KalTA4) zygotes are used for microinjection.
References
Abremski K, Hoess R, Sternberg N (1983) Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32(4):1301–1311
Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K (2008) Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A 105(4):1255–1260. doi:10.1073/pnas.0704963105
Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, Raz E (2005) Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci 118(Pt 17):4027–4038. doi:10.1242/jcs.02522
Braat AK, van de Water S, Goos H, Bogerd J, Zivkovic D (2000) Vasa protein expression and localization in the zebrafish. Mech Dev 95(1-2):271–274
Branda CS, Dymecki SM (2004) Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6(1):7–28
Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M (1989) An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol 209(3):423–432
Ciruna B, Weidinger G, Knaut H, Thisse B, Thisse C, Raz E, Schier AF (2002) Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc Natl Acad Sci U S A 99(23):14919–14924. doi:10.1073/pnas.222459999
Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ (2007) Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol 304(2):811–824. doi:10.1016/j.ydbio.2007.01.033
Distel M, Wullimann MF, Koster RW (2009) Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci U S A 106(32):13365–13370. doi:10.1073/pnas.0903060106
Fan L, Moon J, Wong TT, Crodian J, Collodi P (2008) Zebrafish primordial germ cell cultures derived from vasa::RFP transgenic embryos. Stem Cells Dev 17(3):585–597. doi:10.1089/scd.2007.0178
Gallardo T, Shirley L, John GB, Castrillon DH (2007) Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45(6):413–417. doi:10.1002/dvg.20310
Giniger E, Varnum SM, Ptashne M (1985) Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40(4):767–774
Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265(5168):103–106
Guarente L, Yocum RR, Gifford P (1982) A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A 79(23):7410–7414
Hammond SS, Matin A (2009) Tools for the genetic analysis of germ cells. Genesis 47(9):617–627. doi:10.1002/dvg.20539
Hoess RH, Abremski K (1985) Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 181(3):351–362
Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77(1):61–68
John GB, Gallardo TD, Shirley LJ, Castrillon DH (2008) Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol 321(1):197–204. doi:10.1016/j.ydbio.2008.06.017
Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R (2007) RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131(7):1273–1286. doi:10.1016/j.cell.2007.11.034
Ketting RF (2007) A dead end for microRNAs. Cell 131(7):1226–1227. doi:10.1016/j.cell.2007.12.004
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203(3):253–310. doi:10.1002/aja.1002030302
Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C (2002) An evolutionary conserved region in the vasa 3'UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol 12(6):454–466
Koprunner M, Thisse C, Thisse B, Raz E (2001) A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 15(21):2877–2885. doi:10.1101/gad.212401
Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M (2011) Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 138(22):4831–4841. doi:10.1242/dev.072587dev.072587
Krovel AV, Olsen LC (2002) Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech Dev 116(1–2):141–150
Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236(11):3088–3099
Lakso M, Sauer B, Mosinger B Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H (1992) Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci U S A 89(14):6232–6236
Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT (2005) Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 102(17):6068–6073. doi:10.1073/pnas.0408708102
Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI (2007) Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A 104(22):9410–9415. doi:10.1073/pnas.0611302104
Liu W, Collodi P (2010) Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. FASEB J 24(8):2641–2650. doi:10.1096/fj.09-148403
Liu WY, Wang Y, Sun YH, Wang YP, Chen SP, Zhu ZY (2005) Efficient RNA interference in zebrafish embryos using siRNA synthesized with SP6 RNA polymerase. Dev Growth Differ 47(5):323–331. doi:10.1111/j.1440-169X.2005.00807.x
Liu WY, Wang Y, Qin Y, Wang YP, Zhu ZY (2007) Site-directed gene integration in transgenic zebrafish mediated by cre recombinase using a combination of mutant lox sites. Mar Biotechnol (NY) 9(4):420–428. doi:10.1007/s10126-007-9000-x
Liu X, Li Z, Emelyanov A, Parinov S, Gong Z (2008) Generation of oocyte-specifically expressed cre transgenic zebrafish for female germline excision of loxP-flanked transgene. Dev Dyn 237(10):2955–2962. doi:10.1002/dvdy.21701
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. doi:10.1006/meth.2001.1262S1046-2023(01)91262-9
Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A (2000) Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26(2):116–117
Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN Jr, McAvoy R, Jiang X, Wu Y, He A, Pei Y, Li Y (2007) 'GM-gene-deletor': fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5(2):263–274. doi:10.1111/j.1467-7652.2006.00237.x
Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436(7048):207–213. doi:10.1038/nature03813
Pan X, Wan H, Chia W, Tong Y, Gong Z (2005) Demonstration of site-directed recombination in transgenic zebrafish using the Cre/loxP system. Transgenic Res 14(2):217–223
Phelps CB, Brand AH (1998) Ectopic gene expression in Drosophila using GAL4 system. Methods 14(4):367–379. doi:10.1006/meth.1998.0592
Saito T, Goto-Kazeto R, Arai K, Yamaha E (2008) Xenogenesis in teleost fish through generation of germ-line chimeras by single primordial germ cell transplantation. Biol Reprod 78(1):159–166. doi:10.1095/biolreprod.107.060038
Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H (2007) Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods 4(4):323–326. doi:10.1038/nmeth1033
Seok SH, Na YR, Han JH, Kim TH, Jung H, Lee BH, Emelyanov A, Parinov S, Park JH (2010) Cre/loxP-regulated transgenic zebrafish model for neural progenitor-specific oncogenic Kras expression. Cancer Sci 101(1):149–154. doi:10.1111/j.1349-7006.2009.01393.x
Shaikh AC, Sadowski PD (1997) The Cre recombinase cleaves the lox site in trans. J Biol Chem 272(9):5695–5702
Siegal ML, Hartl DL (1996) Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144(2):715–726
Slanchev K, Stebler J, de la Cueva-Mendez G, Raz E (2005) Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci U S A 102(11):4074–4079. doi:10.1073/pnas.0407475102
Suzuki H, Tsuda M, Kiso M, Saga Y (2008) Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol 318(1):133–142. doi:10.1016/j.ydbio.2008.03.020
Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C (2004) Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77:505–519
Thummel R, Burket CT, Brewer JL, Sarras MP Jr, Li L, Perry M, McDermott JP, Sauer B, Hyde DR, Godwin AR (2005) Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn 233(4):1366–1377. doi:10.1002/dvdy.20475
Traven A, Jelicic B, Sopta M (2006) Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep 7(5):496–499. doi:10.1038/sj.embor.7400679
Urasaki A, Morvan G, Kawakami K (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174(2):639–649. doi:10.1534/genetics.106.060244
Van Doren M, Williamson AL, Lehmann R (1998) Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8(4):243–246
Vergunst AC, Jansen LE, Hooykaas PJ (1998) Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res 26(11):2729–2734
Wang L, Zhang Y, Zhou T, Fu YF, Du TT, Jin Y, Chen Y, Ren CG, Peng XL, Deng M, Liu TX (2008) Functional characterization of Lmo2-Cre transgenic zebrafish. Dev Dyn 237(8):2139–2146. doi:10.1002/dvdy.21630
Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E (2003) dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol 13(16):1429–1434
Westerfield M (1995) The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). University of Oregon, Eugene
Yoon C, Kawakami K, Hopkins N (1997) Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124(16):3157–3165
Yoshikawa S, Kawakami K, Zhao XC (2008) G2R Cre reporter transgenic zebrafish. Dev Dyn 237(9):2460–2465. doi:10.1002/dvdy.21673
Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A (2005) The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435(7040):360–364. doi:10.1038/nature03595
Zhan H, Gong Z (2010) Delayed and restricted expression of UAS-regulated GFP gene in early transgenic zebrafish embryos by using the GAL4/UAS system. Mar Biotechnol (NY) 12(1):1–7. doi:10.1007/s10126-009-9217-y
Acknowledgments
The authors thank Prof. Erez Raz for providing the kop-EGFP-F-nos1-3’UTR construct and Prof. Chi-Bin Chien for sharing the pDestTol2pA2 vector. This work was supported by the China 973 Project (2010CB126306 and 2012CB944504) and the National Science Fund for Excellent Young Scholars of NSFC (31222052).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary Table 1
Primers used in the present study (DOC 71 kb)
Rights and permissions
About this article
Cite this article
Xiong, F., Wei, ZQ., Zhu, ZY. et al. Targeted Expression in Zebrafish Primordial Germ Cells by Cre/loxP and Gal4/UAS Systems. Mar Biotechnol 15, 526–539 (2013). https://doi.org/10.1007/s10126-013-9505-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-013-9505-4