Abstract
The present work was designed to assess the effects of artificially increased high plasma cortisol levels induced by slow-release cortisol implants on the mRNA abundance of the glucocorticoid receptor (GR) in different organs of Sparus aurata (Gilthead sea bream), as well as to evaluate global transcriptional changes in the liver, using the Aquagenomics S. aurata oligo-nucleotide microarray technology. For that purpose, groups of fish were intraperitoneally injected with implants containing two different concentrations of cortisol (50 or 200 μg/g body weight). Blood and organs were sampled after 7 and 14 days of cortisol implantation. Only fish with 200 μg/g implants exhibited a significant rise in plasma cortisol. Thus, we evaluated the expression of the GR in different organs in these fish 7 and 14 days post-implantation. GR mRNA abundance was upregulated in head kidney and heart of fish at both sampling times. In liver and muscle, GR mRNA abundance was upregulated after 14 days, whereas in gills, the GR mRNA transcript was upregulated earlier, at day 7. These results suggest that increased plasma cortisol induced by a slow-release implant of cortisol mimics the overall effects of stress and affects the expression of GR mRNA in a time- and organ-specific manner. Data obtained with the Aquagenomics S. aurata oligo-nucleotide microarray allowed the identification of a total of 491 cortisol-responsive transcripts and highlight the strong intensity of transcriptional modulation in liver of fish implanted with cortisol after 7 days, in contrast to that observed at day 14. Transcriptional remodeling highlighted a significant activity in carbohydrate metabolism mainly in the gluconeogenic pathway linked to downregulation of inflammatory and immune response processes in implanted fish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In teleost fish, cortisol is the main endogenous glucocorticoid that regulates a broad spectrum of physiological functions and plays an important role in the maintenance of both basal and stress-related homeostasis (Mommsen et al. 1999; Tort 2010). Cortisol triggers the activation of specific intracellular responses through glucocorticoid receptors (GRs). The hormone–receptor complex binds the GRE for transactivation but can also bind with other transcription factors to affect their actions on transcription by inhibiting activation of their response elements (transrepression) (Stolte et al. 2006). In different fish species, several isoforms of the cortisol receptor have been described. Cyprinus carpio express three GRs (Stolte et al. 2008), Oncorhynchus mykiss express two (Bury et al. 2003), and Danio rerio and Sparus aurata (Gilthead sea bream) express only one GR (Alsop and Vijayan 2009; Acerete et al. 2007). In S. aurata, the GR is constitutively expressed in different organs, such as the brain, spleen, or gills, but its expression is also regulated under different conditions, such as treatment with bacterial lipopolysaccharide (LPS) (Acerete et al. 2007).
Increased plasma cortisol levels can modify the GR mRNA abundance in target tissues such as the brain, liver, or head kidney in fish. However, divergent findings concerning this subject have been reported (Vijayan et al. 2003; Terova et al. 2005; Aluru and Vijayan 2007, 2009). Despite the available literature, the data concerning GR mRNA abundance in fish consist of snapshots of a given moment and one particular target organ. However, an extensive analysis of GR mRNA abundance in different organs of fish with artificially induced high cortisol levels has not yet been carried out.
In fish, plasma cortisol increase is typically correlated to an increase in secondary physiological indicators of stress, such as plasma glucose (Martínez-Porchas et al. 2009), and changes in these parameters can mirror alterations in metabolic and physiological processes in the whole organism. Thus, to establish a more complete stress profile of the experimental organism not only cortisol but also other indicators of stress more closely related to a specific physiological function, such as plasma glucose, lactate and complement activity were carried out in the current study. Complement activity is a nonspecific defense mechanism in fish and has been described as a good indicator for immunocompetence in S. aurata where complement activity displayed an inverse relationship with high plasma cortisol levels induced by several stressors or exogenous cortisol administration (Tort et al. 1996; Montero et al. 1998; Ortuño et al. 2002).
Keeping in mind previous findings, in the present work we aimed to assess the effects of high plasma cortisol levels artificially induced by cortisol implants on GR mRNA abundance in different organs of S. aurata, including the gills, intestine, spleen, brain, head kidney, heart, liver, and muscle. The gills and intestine are primary sites of cortisol action, with important roles in osmoregulation and mucosal immunity. The spleen is one of the main secondary lymphoid organs for antigen trapping and processing, and the brain is involved in cortisol release and regulation through the stimulation of the hypothalamo–pituitary–interrenal (HPI) axis after the perception of an external stimulus. The fish head kidney/pronephros is a central element of the HPI axis and the major source of corticosteroids. Finally, the liver and muscle represent the majority of body weight and are relevant tissues in which most energetic and intermediary metabolism takes place. In order to evaluate transcriptional changes induced by cortisol treatment in the liver, we used the Aquagenomics S. aurata oligo-nucleotide microarray (SAQ) developed and validated by Boltaña et al. 2011 (unpublished). Our aim was to identify key transcripts to be used in future studies as stress biomarkers.
Material and Methods
Test Animals
S. aurata specimens (108 ± 4.3 g mean weight) were obtained from a local fish farm (Cripesa, L’Ametlla de Mar, Spain). Fish were acclimatized to laboratory conditions for 15 days, maintained in a closed seawater recirculation system, at 15 °C, in a 12-h light/12-h dark cycle and fed a maintenance diet of commercial pellets once a day. Fish were fasted 24 h before the experimental procedures. Water quality indicators, such as dissolved oxygen, pH, nitrite, and ammonia, were analyzed periodically, and the measured values were acceptable considering the particular requirements of this species (Roncarati et al. 2006). All experimental procedures agree with the “International Guiding Principles for Biomedical Research Involving Animals,” and they were supervised by the ethical committee of the Universitat Autònoma de Barcelona.
Experimental Design and Sampling Procedure
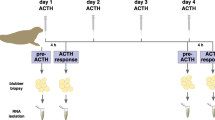
Three experimental groups of six fish were anesthetized with 0.1 g/L of tricaine methane sulphonate (MS222, Sigma-Aldrich E10521). MS222 were dissolved in water taken from the holding aquarium, and fish were introduced into the anesthetic bath. Fish were intraperitoneally (ip) injected with coconut oil implants. This protocol has been shown to produce a slow release of cortisol into the circulation, and the time and dose of cortisol used in the present experiment were chosen based on results previously obtained in S. aurata (Laiz-Carrión et al. 2003). Furthermore, we carried out an in-house preliminary experiment where plasma cortisol levels were increased after implantation with 50, 100, and 200 μg/g body weight of cortisol (Teles et al., unpublished data). Briefly, cortisol implants were prepared by warming the coconut oil at 30 °C and mixing the desired amount of cortisol (hydrocortisone hemisuccinate, Sigma). Fish were ip injected with the implants (5 μl/g body weight) of coconut oil alone (controls) or with the implants containing two different concentrations of cortisol (50 or 200 μg/g body weight). One extra group of fish without the implant (untreated fish) was maintained under the same conditions and sampled at day 0. After 7 and 14 days of cortisol implantation, fish were randomly sampled from duplicate tanks used for each experimental group and sacrificed by over-anesthetization in MS222 (1 g/L). Blood was collected with heparinized syringes from the caudal vein and used to extract plasma using an Eppendorf centrifuge (17,000×g). Blood collection lasted less than 3 min in order to avoid cortisol rise induced by the manipulation during sampling. The spleen, brain, head kidney, heart, liver, muscle, gills, and intestine were carefully dissected out and immediately frozen in liquid nitrogen. Samples were stored at −80 °C for further assays.
Plasma Physiological Measurements
Cortisol was measured using a diagnostic ELISA direct immunoenzymatic method (Immulite 1000 Cortisol, Siemens Healthcare Diagnostics, UK). Glucose and lactate were determined by enzymatic colorimetric analysis in ELISA plates using commercial kits (Biomérieux, France). Plasma osmolality was assessed by measuring freezing point in an osmometer (Osmomat 030 Gonotec, Berlin). Alternative complement pathway activity was determined following the technique described by Sunyer and Tort (1995) with minor modifications for ELISA plates and expressed as titers causing 50 % of hemolysis of rabbit blood cells (ACH50 units).
RNA Isolation and Complementary DNA Synthesis
Total RNA was individually extracted from tissues using 1 ml per sample of TRI reagent (Molecular Research Center) and following the manufacturer's instructions. RNA quantification was carried out with a NanoDrop ND-1000 (Thermo Scientific) and the quality of the RNA checked with the Bioanalyzer 2100 using the 6000 Nano LabChip kit (Agilent Technologies). All RNA integrity number values obtained were >8, indicative of excellent RNA integrity and quality. Four micrograms of RNA was used to synthesize complementary DNA (cDNA) with Superscript III™ reverse transcriptase (Invitrogen) and oligo-dT primer (Promega).
RNA Labeling and Hybridization
A complete description of the microarray platform used in the present research as well as the data presented in this manuscript is accessible through the public Gene Expression Omnibus (GEO) depositories (accession number GSE30782). Briefly, the SAQ oligonucleotide array was designed using eArray (Agilent Technologies) and contains 7,285 transcripts with annotated sequences that were spotted in triplicate (total probes, 21,855), as well as 8,377 expressed sequence tags (ESTs) without annotation and 183 enriched control sequences with 15 replicated probes (total probes, 2,745), and finally 1,417 internal control Agilent probes (N = 43,398). Sequences present on the array were obtained from public resources and an unpublished EST collection (Aquagenomics Consortium, Spain). For microarray analysis, we used livers of S. aurata (N = 18), previously ip injected with 200 μg/g body weight of cortisol and sampled after 7 and 14 days of implantation (n = 6 for each condition). RNA samples were grouped into three pools of two fish thus obtaining a total of three pools per condition. Denatured pooled samples of RNA were reversed transcribed and indirectly labeled with Cy3. RNA labeling, hybridizations, and scanning were performed according to the manufacturer’s instructions. Briefly, total RNA (500 ng) was amplified and Cy3-labeled with Agilent’s One-Color Microarray-Based Gene Expression Analysis (Quick Amp Labeling kit) along with Agilent’s One-Color RNA Spike-In Kit. Each sample (1.65 μg of RNA) was hybridized to S. aurata array (ID 024502, Agilent) at 65 °C for 17 h using Agilent’s GE Hybridisation Kit. Washes were conducted as recommended by the manufacturer using Agilent’s Gene Expression Wash Pack with stabilization and drying solution. Microarrays slides were scanned with Agilent Technologies Scanner model G2505B. Spot intensities and other quality control features were extracted with Agilent’s Feature Extraction software version 10.4.0.0. One-channel TIFF images were imported into the GeneSpring software GX 11.0. After percentile normalization, all data were filtered by comparison of the standard deviation expression among groups (filter by expression). The entities that had values lesser or greater than the standard deviation value were retained. This filter procedure allowed selected samples that have outlier entities, and filters out probes that have a high/low variation expression values across the samples.
A loop design was used for the study, where all RNA samples were labeled with a single color dye (Cy3), and each experimental sample was compared to the control sample (pool without stimulation). This approach allowed us to identify differentially expressed transcripts under three distinct conditions: (1) control and 7-day post-implanted fish, (2) 7 and 14 days post-implantation, and (3) control and cortisol implantation (time independent).
Primer Design, Quantitative Real-Time Polymerase Chain Reaction Assay, and Microarray Validation
The primers used for the GR mRNA abundance study and to validate the microarrays results were designed with Primer3 version 4.0 based on target sequences obtained from S. aurata database (Table 1). To ensure that the pair of primers was specifically amplifying the target sequence, we validated both primer sequences on opposing strands of the GR using the NCBI GenBank database and BLAST software (http://www.ncbi.nlm.nih.gov/BLAST). For quantitative real-time polymerase chain reaction (qPCR) assays, a total volume of 20 μl was used, and every reaction contained 500 nM of each amplification primer, 10 μl of iQ™ SYBR Green Supermix (Bio-Rad), and 5 μl of cDNA. The cDNA dilutions were 1:50 for target genes and 1:500 for the housekeeping gene, 18S. Controls lacking cDNA and controls containing RNA were included. Reactions were run in the iCycler iQ™ Real-time PCR Detection System (Bio-Rad Laboratories), under the protocol: 1 cycle of 95 °C for 5 min, 40 cycles of 95 °C for 10 s, and 60 °C for 30 s, 1 cycle of 95 °C for 1 min, 1 cycle of 55 °C for 1 min, and a melting curve of 81 cycles (from 55 to 95 °C). All samples were run in triplicate. Values for each sample were expressed as “fold differences,” calculated relative to untreated controls and normalized to 18S.
Data Analysis
The physiological parameters data and GR mRNA transcription data were first tested for normality and homogeneity of variance to meet statistical demands. Differences between control and exposed groups were tested using one-way ANOVA, followed by LSD post hoc test, and the significance level was ascertained at P < 0.05. Differences between different exposure times and the same treatment were tested using the two-tailed Student’s t test. Experiments were carried out using test groups of six fish (n = 6). For the microarray data, statistical tests were implemented using the GeneSpring software GX 11.0 to select differentially expressed transcripts (P < 0.01) between control and treatments. One-way ANOVAs were used to explore differences in the transcriptomic profile between treatments. The complete design has been submitted to GEO database with the number GPL13442. Statistica (StatSoft Inc., Tulsa, OK) and SPSS 17.0 programs were used for statistical analyses.
Results
Physiological Response and GR mRNA Abundance
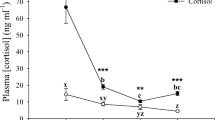
Both cortisol-implanted groups (50 and 200 μg/g) showed elevated plasma cortisol levels. However, this increase was statistically significant only in fish implanted with the higher cortisol concentration after 7 and 14 days of implantation. Cortisol increase at day 14 was significantly lower than at day 7 ( Fig. 1a). Glucose levels were also significantly higher in the plasma of fish with 200 μg/g cortisol implants after 7 days when compared to control and to 50 μg/g implanted fish ( Fig. 1b). Both concentrations of cortisol caused a significant increase in plasma lactate at day 7 after implantation (Fig. 1c), but lactate returned to control levels at day 14. Plasma osmolality was unaltered throughout the experiment (Fig. 1d). Plasma complement activity (ACH50) showed a tendency to decrease in all implanted groups, and was statistically significant 14 days after implantation with both 50 and 200 μg/g cortisol implants (Fig. 1e).
Plasma cortisol (a), glucose (b), lactate (c), osmolality (d), and complement activity ACH50 (e) of S. aurata 7 and 14 days after ip implantation of 5 μl/g body weight (bw) of coconut oil alone (control) and containing 50 or 200 μg/g bw cortisol. Values represent the means and SE (n = 6 per group). Significant differences are highlighted as a versus control at 7 and 14 days, b vs. 50 μg/g at 7 or 14 days, and c vs. 7 days same dose (p < 0.05)
For GR mRNA abundance in distinct organs, when compared to control, 7 days after implantation (200 μg/g cortisol) GR was upregulated in the head kidney [2.6-fold change (FC)], heart, and gills (2.8- and 2.2-fold change, respectively). After 14 days, GR mRNA remained upregulated in the head kidney (2.6-fold change) and heart (2.5-fold change) but was also detected in the liver and muscle (3.9- and 4.7-fold change, respectively). GR expression in the intestine showed a tendency towards upregulation at day 7, though not significant, and was significantly downregulated (0.5-fold change) 14 days after cortisol implantation compared to day 7 implanted group. Additionally, GR mRNA abundance was upregulated in liver (2.6-fold change) and muscle (twofold change) when compared to the day 7 group, and significantly increased in gills at day 7 (2.2-fold change) when compared to the 14-day group. GR mRNA abundance was unaltered in the spleen and brain (Fig. 2).
Gene expression of glucocorticoid receptor (GR) in different tissues of S. aurata 7 and 14 days after ip implantation of 5 μl/g body weight (bw) of coconut oil alone (control) or containing 200 μg/g bw cortisol. Gene expression was analyzed by real-time PCR and normalized to 18S rRNA. Values represent the means and SE (n = 5 per group). Differences between groups are: a vs. control; b vs. 7 days (p < 0.05)
Transcriptional Response
The Aquagenomics S. aurata oligo-nucleotide microarray (SAQ) was used to explore changes in global gene expression in the liver of S. aurata with 200 μg/g body weight cortisol implants over a period of 1 and 2 weeks post-implant. Transcriptomic profiles obtained were significantly different between individuals treated with equal concentrations of cortisol implants in both transcript number and intensity (Fig. 3). A total number of 491 transcripts were differentially expressed after cortisol treatment (7 and 14 days) (Electronic Supplementary Material 1). From these transcripts, 277 (56 %, p < 0.01) were exclusive to fish implanted with cortisol after 7 days when compared to the control group (implanted with coconut oil alone), and 106 of those transcripts were regulated at a FC > 2 (22 % of the total number of transcripts). Among the 277 transcripts, 161 (58 %) were upregulated and 116 (42 %) were downregulated. When comparing day 14 to day 7, we identified 192 transcripts (39 %, p < 0.01) that were exclusive to this time period. A total of 46 transcripts had a FC > 2, which corresponds to 9 % of the total number of transcripts (491). From the 192 transcripts exclusive to day 14, 94 (49 %) were upregulated and 98 (51 %) were downregulated. Twenty-two transcripts were treatment specific and time independent, i.e., common to both sampling times, and from these transcripts, only seven, which represents 1.4 % of the total number of differentially expressed transcripts (491), had a FC > 2.
Venn diagram representing mRNA transcripts differentially expressed (GDE) in the liver of S. aurata treated with cortisol implants (200 μg/g bw). The area of the circles is scaled to the number of transcripts (one-way ANOVA, p < 0.01) and the fold change (FC > 2) expressed for each condition. White circle: 277 transcripts differentially expressed 7 days after cortisol implantation. Black circle: 192 transcripts differentially expressed 14 days after cortisol implantation when compared to the 7 days implanted group (time-dependent). Grey zone: 22 transcripts differentially expressed common to implanted fish (time-independent)
Injection with 200 μg/g cortisol induced changes in mRNA transcripts mainly related with metabolism, energy, and immune response (Tables 2 and 3). At day 7, transcripts encoding proteins involved in the gluconeogenic pathway were differentially regulated, such as phosphoglucomutase 1 (upregulated). This transcript encodes for an enzyme that catalyzes the interconversion of glucose 1-phosphate into glucose 6-phosphate and vice versa (Golicnik et al. 2009). The enolase 1 alpha mRNA transcript, on the other hand, encodes for an enzyme known to have a role in the glycolytic pathway, converting 2-phosphoglycerate into phosphoenolpyruvate (Kim and Dang 2005) but also responds to stress (Ribas et al. 2004).
Transcripts encoding for ribosomal proteins, such as ribosomal protein L13a and S6 kinase alpha4, were upregulated at day 7. Other transcripts presenting an increased mRNA abundance were aquaporin 12 and death activator CIDE 3 with a FC of 3.36 and 3.13, respectively. The aquaporin 12 mRNA transcript encodes for a protein that facilitates the transport of water and small neutral solutes across cell membranes (Zilli et al. 2011). The cell death activator CIDE3 mRNA encodes for proteins involved in apoptosis and belonging to the cell death-inducing DNA fragmentation factor-like effector family (Liang et al. 2003). Transcripts that encode for proteolytic enzymes, such as cdc48 and pitrilysin metalloproteinase 1, were also upregulated at day 7 after cortisol implantation. The cdc48 protein has a large potential array of cellular functions and is directly linked to ubiquitin-mediated protein degradation (Dargemont and Ossareh-Nazari 2012), and pitrilysin metalloproteinase 1 is an ATP-independent protease. The mRNA abundance of zinc-binding protein 1 was increased. This transcript is related with the induction of metallothioneins and other transcripts involved in metal homeostasis (Bourdineaud et al. 2006).
Overall, a downregulation was observed for immune-related mRNA transcripts in the liver 7 days after 200 μg/kg cortisol implantation. We identified relevant transcripts linked to the expression of proteins responsible for antigen recognition, such as T cell receptor alpha and beta chain, major histocompatibility class (MHC) II antigen beta chain, and MHC class II antigen-associated invariant chain. We also observed a downregulation in the mRNA abundance of complement C9, which encodes for the final component of the complement system (Holland and Lambris 2002). This protein when coupled to the downregulation of the complement component C4, common to both exposure times, suggests cortisol-induced regulation of the complement system. The C4 mRNA transcript encodes for a protein with a central role in the activation of the complement system (Holland and Lambris 2002). Further downregulated transcripts related with the immune function were interleukin-22, interleukin-8-like protein, and CXC chemokine and interferon regulatory factor 1. Interleukin-22 protein was recognized to increase its activity during chronic inflammatory diseases, preventing tissue damage during immune responses (Zenewicz et al. 2007). Interleukin-8-like and CXC chemokine proteins function as chemotactic substances, inducing the recruitment of cells of the immune system to the site of infection (Zhonghua et al. 2008). Finally, the jeltraxin transcript, which encodes for a protein similar to C-reactive P component and serum amyloid P (Tadiso et al. 2011), also showed reduced mRNA abundance.
On day 14, the differentially regulated transcripts presenting a FC > 2 were fewer when compared to day 7. At day 14, we identified an upregulation of glutathione peroxidase 1 mRNA transcript, which encodes a member of the glutathione peroxidase family, with a function in hydrogen peroxide detoxification (Winston 1991). We identified downregulated transcripts such as glucose-6-phosphate dehydrogenase, a transcript that encodes for an important enzyme of the pentose phosphate metabolic pathway (Adem and Ciftci 2012).
Microarray results were validated by quantifying the expression of a number of transcripts by qPCR, using the same RNA pools used in the microarray hybridization. Relative correlation between microarray-based and qPCR expression measured target transcript values registers a positive Pearson correlation coefficient (0.88), and was always significantly positive p < 0.001 (Fig. 4), highlighting the high reproducibility of the S. aurata oligo-nucleotide microarray using an independent expression of measurement method (qPCR).
Comparison between microarray and quantitative real-time PCR (qPCR) results using the Pearson coefficient. Expression values for the 11 differentially expressed genes (alcohol dehydrogenase, MHC class II antigen-associated invariant chain, bloodthirsty, enolase 1 alpha-like, MHC class II antigen beta chain, T cell receptor beta chain, ribosomal protein L13, CIDE3, pitrilysin metalloproteinase 1, cdc48, phosphoglucomutase, and zinc-binding protein 1) were compared between microarray probes and real-time qPCR
Discussion
Physiological Response to Cortisol and GR Regulation
In the present study, we observed that cortisol implants increase plasma cortisol levels in S. aurata at day 7 post-implantation. Measured cortisol levels were situated within the physiological range observed for this fish species after chronic stress situations or after exogenous cortisol administration, later at day 14 plasmatic cortisol levels were lower than at day 7 emulating previous findings (Tort et al. 1996; Laiz-Carrión et al. 2003). The decrease in plasma cortisol levels at day 14 may be due to increased cortisol catabolism and feedback regulation resulting in acclimation to chronic stress. These elevated levels were effective in inducing an increase in plasma glucose and lactate levels at 7 days in cortisol-implanted groups, reflecting the regulatory effect of this hormone on carbohydrate metabolism. After 14 days and concomitant with lower plasma cortisol levels, glucose and lactate returned to control levels, suggesting a tradeoff on the steady state of the metabolic function in correlation with the reduction of the plasmatic cortisol levels.
Under the present experimental conditions, cortisol induced a differential GR mRNA abundance that was both tissue-specific and time-dependent. In gills, GR mRNA abundance increased at day 7 and later at day 14 returned to control levels, a result that correlates to the observed plasma cortisol levels. Yada et al. (2008) observed in rainbow trout that the GR mRNA was upregulated in gills during smoltification, a process marked by stark changes in the osmoregulatory function of the gills during seawater adaptation. In the intestine, GR mRNA abundance was downregulated at day 14 which may be associated to decreased receptor affinity, shortening of receptor half-life, and reducing mRNA abundance. In our study, GR mRNA abundance in the spleen was unaltered in implanted groups similar to that observed in chronically stressed rainbow trout (Yada et al. 2007). In whole brain, the GR mRNA abundance was unaltered throughout the experiment. This may possibly be due to our approach using whole brain samples where small changes in distinct brain areas such as the hypothalamus may be masked.
In the head kidney, GR mRNA abundance was consistently upregulated, reflecting the positive regulation of cortisol receptors caused by high plasma cortisol levels. Previously, Vazzana et al. (2010) reported an increase in GR mRNA abundance in Dicentrarchus labrax head kidney 7 days postcortisol implantation. Similar dynamics of GR mRNA abundance were observed in the heart, which is in concordance with the observed glucocorticoid effect upon cardiovascular mRNA expression of the GR (Kayes-Wandover and White 2000). In the liver and muscle, the GR expression pattern was similar, where the GR transcript was upregulated at day 14. Thus, despite the small observed increase in plasma cortisol at day 14, there appears to be an effect on these organs.
Transcriptional Response
We used the Aquagenomics S. aurata oligo-nucleotide microarray (SAQ) platform to explore changes in global gene expression in the liver of S. aurata induced by cortisol implantation. At day 7 after cortisol implantation, we observed an upregulation of transcripts involved in the gluconeogenic pathway such as phosphoglucomutase 1. This further confirms the gluconeogenic role of cortisol, increasing liver gluconeogenic capacity and leading to elevated glucose production, which is corroborated by the increased plasma cortisol and glucose at day 7. Our results are also in accordance with previous findings showing higher phosphoenolpyruvate carboxykinase (Wiseman et al. 2007) and glucose-6-phosphatase mRNA abundances (Momoda et al. 2007) in the liver of stressed rainbow trout. The present data support the role of cortisol in the molecular regulation of the hyperglycemic response to stress in the fish liver, helping the animal to cope with the stress condition as previously shown by Aluru and Vijayan (2007, 2009).
The upregulation of cdc48 and pitrilysin metalloproteinase 1 mRNA abundance at day 7, together with previous findings where an increased abundance of transcripts related to proteolysis was detected in the liver of stressed fish (Saera-Vila et al. 2005; Wiseman et al. 2007), suggests an activation of protein degradation pathways, supplying amino acids as substrates for gluconeogenesis and allowing the elimination of misfolded proteins induced by cortisol. The concomitant upregulation of ribosomal protein L13a and S6 kinase alpha4 mRNAs indicates increased protein synthesis and cell proliferation. The upregulation of the cell death activator CIDE3 suggests an activation of the apoptotic pathways 7 days after cortisol implantation, as previously suggested (Calduch-Giner et al. 2010).
The liver is a crucial organ in the biotransformation of endogenous steroids and xenobiotics. Under the present conditions, we observed a downregulation of the alcohol dehydrogenase transcript at day 7, supporting the idea that glucocorticoids can negatively modulate the biotransformation processes and confirming previous results in the same or other fish species (Aluru and Vijayan 2007; Calduch-Giner et al. 2010). At day 14, glutathione peroxidase 1 mRNA abundance was increased, suggesting increased levels of prooxidants which may or not lead to peroxidation and increased antioxidant defenses.
Stress most often induces immune-suppressive effects in fish, while the cortisol itself is rather an immune modulator (Tort 2011). Under the present conditions, at day 7 after cortisol implantation, we observed a downregulation of immune-related transcripts, such as the T cell receptor and MHC class II. This supports the idea that chronic treatment with cortisol has immune-suppressive effects. We also observed decreased mRNA abundance of transcripts encoding components of the complement pathway, such as complement C9 (at day 7) and C4 (at days 7 and 14). These changes in specific mRNA abundance correlate to the plasma data obtained where a decrease in complement activity was observed. Thus, sustained increases in plasma cortisol may affect susceptibility to disease by suppressing the complement system. Additionally, downregulation of transcripts encoding for inflammatory regulators, namely interleukin-8-like protein, interleukin-22 and CXC chemokine and jeltraxin precursor mRNA were observed. Previous results in rainbow trout macrophages showed that cortisol treatment significantly suppressed LPS-induced transcripts related to the inflammatory response as TNF-∝ and IL-1β (Mackenzie et al. 2006). Thus, cortisol appears to specifically suppress pro-inflammatory signaling pathways.
Globally, we have observed that increased plasma cortisol levels affect GR mRNA abundance differently, and this is likely related to tissue function. Interestingly, the most susceptible tissues, the head kidney, gills, and heart, all play an important role in the regulation of immunity in teleost fish (Acerete et al. 2007; Aluru and Vijayan 2007, 2009). On the other hand, tissues more directly related to metabolic functions, the liver and muscle, appear more refractory to acute changes in plasma cortisol concentrations. Taken together with the data obtained from the liver transcriptome and in the absence of additional environmental stimuli, we suggest that cortisol may act to acutely regulate immunity by suppressing specific immune functions, i.e., complement activity in a tissue-specific manner. On the other hand, cortisol appears to shift metabolic function toward increased catabolic activity reflected by increased plasma glucose. It will be of interest to further study the dynamics of cortisol action upon tissue-specific transcriptional regulation to gain further insight into the regulation of the stress response and its impact upon physiological function.
In conclusion, cortisol implants were effective in inducing artificially high plasma cortisol levels in S. aurata, simulating a chronic stress situation, which is supported by significant changes in the secondary stress indicators. Our results also demonstrate tissue-specific GR mRNA abundance associated with cortisol levels and the exposure time. The results obtained with the Aquagenomics S. aurata oligo-nucleotide microarray platform highlight the strong intensity of transcriptional modulations in the liver of fish implanted with cortisol during 7 days, in contrast to the observed at day 14. Cortisol induced regulation of transcripts involved in different biological functions, such as gluconeogenesis and glycogenolysis, inflammation and immune system, proteolysis and xenobiotic metabolism. The present results contribute to improving knowledge of the transcriptomic response to stress in this commercially important species. This may lead to the identification of candidate transcripts that can be used as potential new biomarkers in future studies and to uncover the underpinning molecular mechanisms in the liver relevant to the stress response.
References
Acerete L, Balasch JC, Castellana B, Redruello B, Roher N, Canario AV, Planas JV, MacKenzie S, Tort L (2007) Cloning of the glucocorticoid receptor (GR) in gilthead sea bream (Sparus aurata) differential expression of GR and immune genes in gilthead sea bream after an immune challenge. Comp Biochem Physiol B 148:32–43
Adem S, Ciftci M (2012) Purification of rat kidney glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and glutathione reductase enzymes using 2′,5′-ADP Sepharose 4B affinity in a single chromatography step. Protein Expres Purif 81:1–4
Alsop D, Vijayan MM (2009) The zebrafish stress axis. Molecular fallout from the telesot specific genome duplication event. Gen Comp Endocrinol 16:62–66
Aluru N, Vijayan MM (2007) Hepatic transcriptome response to glucocorticoid receptor activation in rainbow trout. Physiol Genom 31:483–491
Aluru N, Vijayan MM (2009) Stress transcriptomics in fish: a role for genomic cortisol signaling. Gen Comp Endocrinol 164:142–150
Bourdineaud JP, Baudrimont M, Gonzalez P, Moreau JL (2006) Challenging the model for induction of metallothionein gene expression. Biochimie 88:1787–1792
Bury NR, Sturm A, Le Rouzic P, Lethimonier C, Ducouret B, Guiguen Y, Robinson-Rechavi M, Laudet V, Rafestin-Oblin ME, Prunet P (2003) Evidence for two distinct functional glucocorticoid receptors in teleost fish. J Mol Endocrinol 31:141–156
Calduch-Giner JA, Davey G, Saera-Vila A, Houeix B, Talbot A, Prunet P, Cairns MT, Pérez-Sánchez J (2010) Use of microarray technology to assess the time course of liver stress response after confinement exposure in gilthead sea bream (Sparus aurata L.). BMC Genom 11:193, http://www.biomedcentral.com/1471-2164/11/193
Dargemont C, Ossareh-Nazari B (2012) Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim Biophys Acta 1823:138–144
Golicnik M, Olguin LF, Feng G, Baxter NJ, Waltho JP, Williams NH, Hollfelder F (2009) Kinetic analysis of beta-phosphoglucomutase and its inhibition by magnesium fluoride. J Am Chem Soc 131:1575–1588
Holland MCH, Lambris J (2002) The complement system in teleosts. Fish Shellfish Immunol 12:399–420
Kayes-Wandover KM, White PC (2000) Steroidogenic enzyme gene expression in the human heart. J Clin Endocrinol Metab 85:2519–2525
Kim JW, Dang CV (2005) Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30:142–150
Laiz-Carrión R, Rio MPM, Miguez JM, Mancera JM, Soengas JL (2003) Influence of cortisol on osmoregulation and energy metabolism in gilthead sea bream Sparus aurata. J Exp Zool 298A:105–118
Liang L, Zhao M, Xu Z, Yokoyama KK, Li T (2003) Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem J 370:195–203
MacKenzie S, Iliev D, Liarte C, Koskinen H, Planas JV, Goetz FW, Mölsä H, Krasnov A, Tort L (2006) Transcriptional analysis of LPS-stimulated activation of trout (Oncorhynchus mykiss) monocyte/macrophage cells in primary culture treated with cortisol. Mol Immunol 43:1340–1348
Martínez-Porchas M, Martínez-Córdova LR, Ramos-Enriquez R (2009) Cortisol and glucose: reliable indicators of fish stress? Pan-Am J Aquat Sci 4:158–178
Mommsen TP, Vijayan MM, Moon TW (1999) Cortisol in teleosts: dynamics, mechanisms of action, and metabolic regulation. Rev Fish Biol Fisheries 9:211–268
Momoda TS, Schwindt AR, Feist GW, Gerwick L, Bayne CJ, Schreck CB (2007) Gene expression in the liver of rainbow trout, Oncorhynchus mykiss, during the stress response. Comp Biochem Physiol D 2:303–315
Montero D, Tort L, Izquierdo MS, Robaina L, Vergara JM (1998) Depletion of serum alternative complement pathway activity in gilthead seabream caused by α-tocopherol and n-3 HUFA dietary deficiencies. Fish Physiol Biochem 18:399–407
Ortuño J, Esteban A, Meseguer J (2002) Lack of effect of combining different stressors on innate immune responses of sea bream (Sparus aurata L.). Vet Immunol Immunopathol 84:17–27
Ribas L, Planas JV, Barton B, Monetti C, Bernadini G, Saroglia M, Tort L, MacKenzie S (2004) A differentially expressed enolase gene isolated from the gilthead sea bream (Sparus aurata) under high-density conditions is up-regulated in brain after in vivo lipopolysaccharide challenge. Aquaculture 241:195–206
Roncarati A, Melotti P, Dees A, Mordenti O, Angellotti L (2006) Welfare status of cultured seabass (Dicentrarchus labrax L.) and seabream (Sparus aurata L.) assessed by blood parameters and tissue characteristics. J Appl Ichthyol 22:225–234
Saera-Vila A, Calduch-Giner JA, Gómez-Requeni P, Médale F, Kaushik S, Pérez-Sánchez J (2005) Molecular characterization of gilthead sea bream (Sparus aurata) lipoprotein lipase. Transcriptional regulation by season and nutritional condition in skeletal muscle and fat storage tissues. Comp Biochem Physiol B 142:224–232
Stolte EH, van Kemenade BMLV, Savelkoul HFJ, Flik G (2006) Evolution of glucocorticoid receptors with different glucocorticoid sensitivity. J Endocrinol 190:17–28
Stolte EH, de Mazon AF, Leon-Koosterziel KM, Jesiak M, Bury NR, Sturm A, Savelkoul HF, van Kemenade BM, Flik G (2008) Corticosteroid receptors involved in stress regulation in common carp, Cyprinus carpio. J Endocrinol 198:403–417
Sunyer JO, Tort L (1995) Natural hemolytic and bactericidal activities of sea bream Sparus aurata serum are effected by the alternative complement pathway. Vet Immunol Immunopathol 45:333–345
Tadiso TM, Krasnov A, Skugor S, Afanasyev S, Hordvik I, Nilsen F (2011) Gene expression analyses of immune responses in Atlantic salmon during early stages of infection by salmon louse (Lepeophtheirus salmonis) revealed bi-phasic responses coinciding with the copepod-chalimus transition. BMC Genomics 12:141. doi:10.1186/1471-2164-12-141
Terova G, Gornati R, Rimoldi S, Bernardini G, Saroglia M (2005) Quantification of a glucocorticoid receptor in sea bass (Dicentrarchus labrax, L.) reared at high stocking density. Gene 357:144–151
Tort L (2010) Stress in farmed fish. Its consequences in health and performance. In: Koumoundouros G (ed) Recent advances in aquaculture research. Transworld Research Network. ISBN 978-81-7895-7. Trivandrum, Kerala, India
Tort L (2011) Stress and immunomodulation in fish. Develop Comp Immunol 35:1366–1375
Tort L, Gomez E, Montero D, Sunyer JO (1996) Serum haemolytic and agglutinating activity as indicators of fish immunocompetence: their suitability in stress and dietary studies. Aquacult Int 4:31–41
Vazzana M, Vizzini A, Sanfratello MA, Celi M, Salerno G, Parrinello N (2010) Differential expression of two glucocorticoid receptors in seabass (teleost fish) head kidney after exogeneous cortisol inoculation. Comp Biochem Physiol A 157:49–54
Vijayan MM, Raptis S, Sathiyaa R (2003) Cortisol treatment affects glucocorticoid receptor and glucocorticoid-responsive genes in the liver of rainbow trout. Gen Comp Endocrinol 132:256–263
Winston GW (1991) Oxidants and antioxidants in aquatic animals. Comp Biochem Physiol C 100:173–176
Wiseman S, Osachoff H, Bassett E, Malhotra J, Bruno J, VanAggelen G, Mommsen TP, Vijayan MM (2007) Gene expression pattern in the liver during recovery from an acute stressor in rainbow trout. Comp Biochem Physiol D 2:234–244
Yada T, Azuma T, Hyodo S, Hirano T, Grau EG, Schreck CB (2007) Differential expression of corticosteroid receptor genes in rainbow trout (Oncorhynchus mykiss) immune system in response to acute stress. Can J Fish Aquat Sci 64:1382–1389
Yada T, Hyodo S, Schreck CB (2008) Effects of seawater acclimation on mRNA levels of corticosteroid receptor genes in osmoregulatory and immune systems in trout. Gen Comp Endocrinol 156:622–627
Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA (2007) Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity 27:647–659
Zhonghua C, Chunpin G, Yong Z, Kezhi X, Yaou Z (2008) Cloning and bioactivity analysis of a CXC ligand in black seabream Acanthopagrus schlegeli: the evolutionary clues of ELR + CXC chemokines. BMC Immunol 9:66. doi:10.1186/1471-2172-9-66
Zilli L, Beirão J, Schiavone R, Herraez MP, Cabrita E, Storelli C, Vilella S (2011) Aquaporin inhibition changes protein phosphorylation pattern following sperm motility activation in fish. Theriogenology 76:737–744
Acknowledgments
This work was supported by the Plan Nacional de Investigación (BFU2009-07354, and Consolider-Aquagenomics (CSD 2007-00002), Government of Spain. Financial support to Mariana Teles provided by FCT (SFRH/BPD/34449/2006, Government of Portugal) and (JCI-2010-06509 Government of Spain) is thankfully acknowledged. Thanks are also given to Roger Traveset, Pilar Tudela, and Neus Riuet for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Teles, M., Boltaña, S., Reyes-López, F. et al. Effects of Chronic Cortisol Administration on Global Expression of GR and the Liver Transcriptome in Sparus aurata . Mar Biotechnol 15, 104–114 (2013). https://doi.org/10.1007/s10126-012-9467-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-012-9467-y