Abstract
The females and unexpected males of gynogenetic red crucian carps (GRCC) with the 1:1 sex ratio were found in the progeny of the distant crossing of red crucian carp (RCC; ♀, 2n = 100) × blunt snout bream (BSB; ♂, 2n = 48). The females and males of GRCC were fertile, and they mated each other to generate the red crucian carps (GRCC1) and another variational gray crucian carps (GGCC). The GRCC and their offspring were proved to be diploids (2n = 100) with one to three microchromosomes by examining the chromosomal metaphases. The evidences for the male’s genetic effect in GRCC were provided by means of fluorescence in situ hybridization, Sox-HMG DNA markers, and microsatellite DNA markers. The genotypic variances of GRCC resulted in their phenotypic variances which were quite different from their maternal parent. It was concluded that the formation of the male gynogenetic fish in GRCC resulted from the genetic leakage of the paternal fish in the form of the microchromosomes including the paternal male-determining gene. After being activated by the sperm of BSB, which was inactivated and finally degraded but left the microchromosomes, the egg of RCC, in which the 50 chromosomes were spontaneously doubled to 100 chromosomes, developed into the diploid male gynogenetic fish. The formation of the bisexual GRCC and their progeny indicated that the distant hybridization could generate the bisexual diploid gynogenetic fish with genetic variation derived from the paternal fish, which is of great significance in both fish genetic breeding and evolutionary biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More and more examples suggested that hybridization seems to facilitate speciation and adaptive radiation in animals and plants (Mallet, 2007). By means of distant hybridization, it is available to form the polyploid population, for example, the bisexual allotetraploid hybrids of red crucian carp × blunt snout bream (Liu et al. 2007b), and the bisexual allotetraploid hybrids of red crucian carp × common carp (Liu et al. 2001; Liu et al. 2007a). However, there is rare report on the formation of the bisexual gynogenetic fish by hybridization, which is potential to become new fish population. In nature, there existed a kind of fish (Poecilia formosa) which reproduced by gynogenesis and produced all-female offspring. Nevertheless, by accident, the natural gynogenetic male of this species was found. Unfortunately, it was quite difficult to confirm what the paternal fish of this male natural gynogenetic fish was. It was concluded that this species was of hybrid origin (Hubbs and Drewry 1959). Further studies (Schartl et al. 1995; Nanda et al. 2007) indicated that the males of this kind of gynogenetic species resulted from the microchromosomes derived from the related bisexual host species. It was concluded the formation of the microchromosomes was probably due to the interspecific hybridization with closely related species (Lamatsch et al. 2004). However, it was unclear what the real sperm donor for this gynogenetic species was. If we clearly know the paternal fish of the gynogenetic fish, it favors us to analyze the genetic relationship between the gynogenetic fish and their parents.
Distant hybridization is defined as the interspecific specific crossing. It is a useful strategy for the hybrid offspring to change in genotypes and phenotypes. Interspecific hybridization normally results in genome-level alterations including the occurrence of triploid hybrids and tetraploids and subgenome-level alterations such as the formation of the microchromosomes. In the catalog, red crucian carp (Carassius auratus red var.; RCC) with 100 chromosomes and blunt snout bream (Megalobrama amblycephala; BSB) with 48 chromosomes belong to different subfamilies. RCC falls into the Cyprininae subfamily, while BSB is attributed to the Cultrinae subfamily (Yu, 1989). Both RCC and BSB are diploid bisexual species. In feeding habit, RCC is omnivorous, whereas BSB is herbivore.
Artificial gynogenesis, an induced developmental process in which the maternal genome is activated by genetically inactivated sperm, has been successfully made in many fishes, such as the red crucian carp (C. auratus red var.; Sun et al. 2007), Japanese crucian carp (Carassius cuvieri; Sun et al. 2006), Half-Smooth Tongue Sole (Cynoglossus semilaevis; Chen et al. 2009a), and Large Yellow Croaker (Pseudosciaena crocea; Li et al. 2008)). In our previous study (Sun et al. 2007), we made the artificial gynogenesis of RCC in which the UV-treated sterile sperm of BBS was used to activate the eggs of RCC to develop following the cold shock (0–4°C) for 30 min to double the eggs’ chromosomes. In that study, we only obtained all-female gynogenetic progeny, no male being found. In this study, with the same parents, RCC and BSB, we obtained both males and females of the natural gynogenetic RCC, without any artificial treatment to the paternal sperm and maternal eggs, suggesting that the distant hybridization is beneficial to the formation of the bisexual gynogenetic fish with the microchromosomes derived from the paternal fish. The main differences between the natural gynogenesis in this study and the artificial gynogenesis are the UV treatment to the sperm of BSB and the cold shock to the eggs of RCC. The UV treatment makes the sperm of BSB genetically sterile. In evolutionary biology, the bisexual fertile gynogenetic species would be advantageous over the mon-sexual fertile gynogenetic species because the former is easier to reproduce than the latter. So the formation of the bisexual fertile gynogenetic red crucian carps (GRCC) and GGCC with genetic variation makes it possible to produce two kinds of new crucian carp populations, which is the first report on the formation of the bisexual fertile diploid gynogenetic fish and is of great significance in evolutionary biology.
Materials and Methods
Animals and Crosses
Blunt snout bream and red crucian carp were obtained from The Protection Station of Polyploidy Fish in Hunan Normal University. During the reproductive seasons (from April to June) in 2006 and 2007, each 15 mature females and 15 mature males of both RCC and BSB were chosen as the maternal fish and paternal fish, respectively. The crossings were performed by two groups. In the first group, RCC was used as the maternal parent, and BSB was used as the paternal fish. In the second one, the maternal fish and paternal fish were reversed. The mature eggs and milt of RCC and BSB were fertilized, and the embryos developed in the culture dishes at the water temperature of 19–20°C. In each cross, 2,000 embryos were taken at random for the examination of the fertilization rate (number of embryos at the stage of gastrula/number of eggs × 100%) and hatching rate (number of hatched fry/number of eggs × 100%). The hatched fry were transferred to the pond for further culture.
Because in the reverse cross-BSB (♀) × RCC (♂) there was no living progeny while in the cross-RCC (♀) × BSB (♂) there existed the living offspring including diploid gynogenetic red crucian carp, triploid and tetraploid hybrids, in the present study, for abbreviation, we referred to diploid gynogenetic red crucian carps as GRCC, triploid hybrids as 3nRB hybrids, and tetraploid hybrids as 4nRB hybrids. We also referred to diploid hybrids (embryos) as 2n RB hybrids.
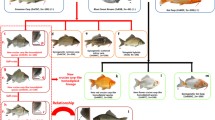
At the age of 3 months, GRCC (F1) were detected by their red body color, while the 3nRB and 4nRB hybrids were gray in the body color. At the age of 1 year, both the mature males and females of GRCC produced the white milt and gray eggs, respectively. The mature milt and eggs of GRCC were striped out and were fertilized to form the living offspring (F2). Two thousand embryos of the offspring were taken at random for the examination of the fertilization rate and hatching rate. The hatched fry were transferred to the pond for further culture. There existed two types of fish in F2 at the age of 3 months, one being red on the body color which was the same as GRCC and was abbreviated as GRCC1, the other being gray on the body color and was abbreviated as GGCC. At the age of 1 year, each 500 GRCC, GRCC1, and GGCC were at random examined for the sex ratios of the females to males. All the mating procedure and the formation of GRCC and the polyploid hybrids as well as GRCC1 and GGCC were illustrated in Fig. 1.
Morphological Traits and Feeding Habit of GRCC and Their Progeny
At the age of 1 year, 20 GRCC, 20 GRCC1, 20 GGCC, 20 RCC, and 20 BSB were morphologically examined, respectively. The examined measurable traits included the average values of the whole length, body length and width, head length and width, and tail length and width. The average ratios of body length to whole length (BL/WL), body width to body length (BW/BL), head length to body length (HL/BL), head width to head length (HW/HL), tail width to tail length (TW/TL), and head width to body width (HW/BW) were calculated. The examined countable traits included the number of dorsal fin, abdomen fin, anal fin, lateral scale, and upper and lower lateral scale. For both measurable and countable data, we used the software of SPSS to analyze the covariance of the data between two kinds of fishes in GRCC, GGCC, GRCC1, RCC, and BSB.
The feeding habit of GRCC, GGCC, GRCC1, and RCC was also investigated.
Examination of the Chromosomal Metaphases
To determine ploidy, chromosome counts were done on the embryos of RCC (♀) × BSB (♂) at the stage of tail bud. Four hundred embryos were treated with 4% Trypsin to get rid of the embryos’ envelope. Then all the samples were grinded in 0.8% NaCl and centrifuged for 1 min at the speed of 1,500 rpm. The 0.5% colchicine was used for 3 h to arrest the chromosomes at metaphase, and the hypotonic treatment was accomplished with 0.075 M KCl at 37°C for 40–60 min, followed by fixation in 3:1 methanol–acetic acid (three changes). Cells were dropped on cold, wet slides, stained for 30–60 min in 4% Giemsa in pH 7.0 phosphate buffer and observed under the light microscope with oil lens.
The chromosomes of the adults of 3nRB (3n = 124) and 4nRB (4n = 148) hybrids have been checked in our previous study (Liu et al. 2007b). To further determine ploidy of the adults of GRCC, GRCC1, and GGCC, chromosome counts were also done on kidney tissue for each ten males and ten females of these three kinds of fish at age of 6 months, respectively. After culture for 1~3 days at the water temperature of 18~22°C, the samples were injected with concanavalin for 1~3 times at a dose of 2∼8 μg/g body weight. The interval time of injection was 12~24 h. Six hours prior to dissecting, each sample was injected with colchicine at a dose of 2∼4 μg/g body weight. The kidney tissue was grinded in 0.9% NaCl, followed by hypotonic treatment and fixation in the same way as described in the chromosome examination for embryos. For each type of fish, 400 metaphase spreads (20 metaphase spreads in each sample) of chromosomes were analyzed. Preparations were examined under the light microscope with oil lens.
The shape and number of chromosomes including the microchromosomes in both embryos and adults were analyzed. Good-quality metaphase spreads were photographed and used for analysis of karyotype. Lengths of entire chromosomes and long and short arms were measured. Chromosomes were classified based on their long-arm to short-arm ratios according to the reported standards (Levan et al. 1964); values of 1.0–1.7 were classified as metacentric (m); 1.7–3.0 as submetacentric (sm), 3.1–7.0 as subtelocentric (st), and 7.1 as telocentric (t) chromosome.
Examination by Fluorescence In Situ Hybridization
The method of fluorescence in situ hybridization (FISH) was used to identify the genetic traits of RCC, BSB, and GRCC. The procedure for the preparation of the chromosome spread for FISH was the same as the above description. Then the hybridization was performed as follows: One pair of primers (5′-TATGCCCGATCTCGTCTGATC-3′ and 5′-CAGGTTGGATGGCCGTAAGC-3′; Masaru and Hideo 1998) was synthesized to amplify the 5S rDNA-related sequences of RCC; the purified polymerase chain reaction (PCR) product labeled with Dig-11-dUTP (Roche) was used as probe. After pretreatment with 2× SSC and 100% ethanol, the slides with chromosome metaphase spreads were denatured in 70% deionized formamide/2× SSC for 2 min at 75°C, dehydrated in a 70% (−20°C) and 100% ethanol series for 5 min each, and then air-dried. One hundred nanograms of labeled probes were blended with 2 μl 20× SSC; 4 μl deionized formamide and 2 μl 50% dextran sulfate were denatured for 5 min in boiling water and then were placed on the slides carrying denatured metaphase chromosomes under a 24 × 50 mm2 coverslip. Hybridization was carried overnight at 37°C in a moist chamber. After a series of post-hybridization washes were performed, the Dig-11-dUTP localization was achieved with 10 μl 1 μg/ml flourescein isothiocyanate-conjugated antidigoxigenin antibody from sheep (Roche). The slides, which were covered in 10 μl antifade solution containing 0.5 μg/ml of 4,6-diamidino-2-phenylindole, were viewed under a Leica inverted DMIRE2 microscope image system (Leica, Germany). Each five males and five females of RCC, BSB, and GRCC, at the age of 6 months, were examined for the hybridizing signal analysis. For each type of fish, 200 metaphase spreads (20 metaphase spreads in each sample) of the chromosomes for hybridizing signal were analyzed.
Observation of Sexes and Gametes
At the age of 6 months, the white semen was stripped out from the males of GRCC, and at the age of 1 year, the mature eggs were stripped out from the females of GRCC. The 4nRB hybrids were fertile at the age of 2 years, whereas the 3nRB hybrids were sterile even at the age of 3 years. The sex ratios of the females to males in GRCC, GRCC1, GGCC, and 4nRB hybrids were recorded.
The semen of GRCC was sucked with a clean sucker and moved into 2.5% glutaraldehyde solution for shape observation. The semen was centrifuged at the speed of 2,000 rpm for 1 min, then was fixed in 4% glutaraldehyde solution for a night, and finally was fixed in 1% osmic acid solution for 2 h. The spermatozoa were observed with a scanning electron microscope (JEOL-6360, made in Japan) after they were treated with alcohol dehydrating, dropping on slides, desiccating, and atomizing gilt. The mature testes of GRCC were fixed in 3% glutaraldehyde solution, then moved to 1% osmic acid solution, and finally embedded in Epon812. The ultrathin sections were cut and stained with uranyl acetate and lead citrate. The electron microscope (JEOL-1230, made in Japan) was used to observe the ultra-structure of the testes.
The mature testes and ovaries of GRCC were moved and fixed in Bouin’s solution for the preparation of tissue sections. The paraffin-embedded sections were cut and stained with hematoxylin and eosin. The structure of the mature testes and ovaries was observed by a light microscope and photographed with a Pixera Pro 600ES.
Examination by the DNA Markers of Sox Genes
Sox genes are characterized by a conserved DNA sequence encoding a high-mobility group (HMG) domain of 80 amino acids, which is responsible for specific DNA sequence binding. The HMG-box DNA fragments belonging to Sox genes were used as the DNA markers to identify the genetic traits of the animal’s samples and were described in our previous study (Chen et al. 2009b). In the present study, the HMG-box DNA fragments were amplified by PCR using the degenerate primers P (+) (5′-TGA A G C G A C C CA T G A A (C/T) G-3′) and P (−) (5′-A G G T C G (A/G) T A C T T (A/G) T A (A/G) T-3′). The genomic DNAs extracted from blood cells of GRCC, GGCC, GRCC1, and RCC by routine approaches (Sambrook et al. 1989) were used as templates. The polymerase chain reaction was performed in a volume of 25 μl with about 80 ng genomic DNA, 1.5 mM of MgCl2, 200 μM of each dNTP, 0.3 μM of each primer, and 0.9 U of Taq polymerase (Takara). The cycling program was 35 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 80 s, with a final extension of 10 min at 72°C. All the PCR products including two DNA fragments in GRCC and GRCC1 and three DNA fragments in GRCC1 and RCC were separated in 1.5% agarose gels.
After electrophoresis, all DNA fragments of PCR products in GRCC, GGCC, GRCC1, and RCC were purified using Gel Extraction Kit (Sangon) and ligated into the pMD-18T vector. The plasmids were amplified in DH5α. The inserted DNA fragments in pMD-18T vector were sequenced by an automated DNA sequencer (ABI PRISM 3730). The sequences were aligned with the corresponding sequences of Sox genes in zebra fish, rainbow trout, medaka, rice field eel, mouse, and common carp etc. derived from NCBI data base using Jellyfish (2.1) software and named accordingly. Sequence homology and variation among the DNA fragments amplified from GRCC, GGCC, GRCC1, and RCC were analyzed using Clustal W software (www.ebi.ac.uk/clustalw/intex.html).
Examination by the Microsatellite DNA Markers
The microsatellite DNA markers which includes much larger amounts of repetitive DNA were widely used as the DNA markers to examine the genetic variances of the samples. In the present study, the total genomic DNA was isolated from the whole blood collected from the caudal vein in ten females of RCC, five males and five females of GRCC, and ten males of BSB, using a reported standard phenol–chloroform procedure (Sambrook et al. 1989). The concentration and quality of DNA were assessed by agarose gel electrophoresis, then samples were stored at 4°C until use.
Five pairs of primers (MFW1, MFW7, MFW15, MFW17, MFW21) were synthesized to the flanking regions including the repeated (CA)n dinucleotide microsatellites according to the published sequences (Crooijmanns et al. 1997). The microsatellite loci were amplified with the above primers in 20 μL reaction volumes: containing 10 mM Tris–HCl, pH 8.8, 50 mM KCl, 1.25 mM MgCl2, 0.15 mM of each dNTP, 0.5 μM of both forward and reverse primers, 50–80 ng of genomic DNA, and 0.5 U Taq polymerase. Amplications were performed in a PCR thermocycler according to the following reaction profile: one cycle at 94°C for 5 min, 30 cycles at 94°C for 30 s, the locus-specific annealing temperature (Tm in Table 1) for 30 s, 72°C for 45 s; and a final extension at 72°C for 10 min. To determine allelic variation at each locus, PCR products were separated on an 8% polyacrylamide gel (32% formamide, 5.6 M urea). PCR products were sized by pBR322 DNA/Mspl ladder.
Results
Morphological Traits and Feeding Habit
The formation procedure of GRCC and their offspring was indicated in Fig. 1. The appearance traits of RCC (Fig. 2a), BSB (Fig. 2b), GRCC (Fig. 2c), GRCC1 (Fig. 2d), and GGCC (Fig. 2e) were shown in Fig. 2. It was easy to distinguish the polyploid hybrids and GRCC. On the body color, the triploid and tetraploid hybrids were gray (Liu et al. 2007b), while GRCC was red which was similar to the body color of RCC. The males and females of GRCC mated to generate GGCC with gray body color and GRCC1 with red body color.
Table 1 presented the examined measurable traits and countable traits in GRCC, GRCC1 GGCC, RCC, and BSB. For the measurable traits, the BL/WL and HL/BL in GRCC and their progeny (GRCC1 and GGCC) were immediate to those of RCC and BSB and were significantly different from those of RCC and BSB. The BW/BL and TW/TL in GRCC and their progeny exceed those in either RCC or BSB and were significantly different from those of RCC and BSB. The HW/BW in GRCC and their progeny was under that in either RCC or BSB and was significantly different from that of RCC or BSB. The HW/HL in GRCC was higher than that in either RCC or BSB. However, the HW/HL in GRCC1 and GGCC was lower or equal to that of BSB and higher that of RCC.
For the countable traits, except for the number of dorsal fins in which GRCC and their progeny was intermediate to RCC and BSB, the other traits including number of lateral scales, number of upper lateral scales, number of lower lateral scales, number of abdominal fins, and number of anal fins in GRCC and their progeny were close to those of RCC and significantly different (P > 0.01) from those of BSB.
Regarding the feeding habit, like BSB, GRCC and their progeny were herbivores.
Chromosomes and Karyotype
Table 2 indicated the distribution of the chromosome number in 400 embryos of RCC (♀) × BSB (♂). The embryos bearing the chromosomes ranging from 70 to 76, in which 73.3% metaphases had 74 chromosomes (Fig. 3a), were diploid hybrids. The embryos with the chromosomes ranking from 98 to 102, in which 82.1% metaphase possessed 100 chromosomes with one to three microchromosomes, were diploid gynogenetic fish. The embryos having chromosomes arraying from 122 to 126, in which 83.9% metaphases showed 124 chromosomes, were triploid hybrids. The embryos possessing chromosomes ranking from 146 to 150, in which 94.4% metaphases presented 148 chromosomes, were tetraploid hybrids. The diploid hybrids, diploid gynogenetic fish, triploid hybrids, and tetraploid hybrids accounted for 7.5%, 14%, 15.5% and 63%, respectively, in all. The chromosomal metaphases of embryos in diploid gynogenetic fish and triploid and tetraploid hybrids were the same as those of their adults which were presented in the present study (GRCC, Fig. 3b) and in our previous study (triploid and tetraploid hybrids, Liu et al. 2007b).
Chromosome spreads at metaphase in diploid hybrid embryo (2nRB), adults of GRCC, GGCC, and GRCC1. a The 74 chromosomes of 2nRB, in which the biggest submetacentric chromosome from BSB was indicated by an arrow, bar = 3 μm; b the 100 chromosomes and three microchromosomes (arrows) of GRCC, bar = 3 μm; c the 100 chromosomes and three microchromosomes (arrows) of GGCC, bar = 3 μm; d the 100 chromosomes and three microchromosomes (arrows) of GRCC1, bar = 3 μm; e the karyotypes of GRCC: 22 m + 34 sm + 22 st + 22 t + 3 microchromosomes (arrow), bar = 3 μm
In the adults of the progeny of the hybrids of RCC (♀) × BSB (♂), the hybrids including the triploid and tetraploid hybrids (Liu et al. 2007b) and the diploid gynogenetic fish (GRCC) were found. But no survival of the adult diploid hybrid with 74 chromosomes was observed. The chromosome number of the adults of GRCC, GRCC1, and GGCC were presented in Table 3. Of all examined samples in GRCC, 95.75% of chromosomal metaphases possessed 100 chromosomes (Table 3). In the 20 examined samples (ten males and ten females), one to three microchromosomes were found in all males (Table 3; Fig. 3b) and in eight out of ten females. The karyotype of GRCC was 22m+34sm+22st+22t+1~3 mc (microchromosomes; Fig. 3e). Except for the microchromosomes, this karyotype was the same as that of the normal red crucian carp as reported in our previous study (Liu et al. 2001). Of all examined examples in GRCC1, 98% of chromosomal metaphase had 100 chromosomes (Table 3). In the 20 examined samples (ten males and ten females), there existed one to three microchromosomes in all males (Fig. 3c) and in seven out of ten females. Their karyotype was the same as that of GRCC. In the examined samples of GGCC, 96.25% of chromosomal metaphases had 100 chromosomes. In the 20 examined samples (ten males and ten females), all the males had one to three microchromosomes, and seven out of ten females possessed one to three microchromosomes. Their karyotype was also the same as that of GRCC.
Fluorescence In Situ Hybridization
The results of FISH (Table 4) showed that there were two strong and one weak hybridizing signals in RCC (Fig. 4a), while there was no hybridizing signal in BSB (Fig. 4b). On the other hand, there were one strong and one weak hybridizing signals in GRCC (Fig. 4c), suggesting that the DNA structures related to 5S rDNA in GRCC were changed. In the ten examined samples of GRCC (five males and five females), all males and three out of five females possessed one to three microchromosomes (Fig. 4c).
Sex Ratio of GRCC and Their Progeny
In the crossing of RCC (♀) × BSB (♂), we observed higher fertilization rate (67%) and hatching rate (58%) and obtained about 100,000 and 150,000 living progeny in 2006 and 2007, respectively. In the reverse crossing RCC (♂) × BSB (♀), there was no survival. In the offspring of RCC (♀) × BSB (♂), the polyploid hybrids including the triploid and tetraploid hybrids with the gray body color accounted for 67%, while GRCC with the red body color accounted for 33%. In 2007, by mating the males and females of GRCC, we obtained 20,000 living offspring with 91% fertilization rate and 75% hatching rate, in which GRCC1 with the red body color and GGCC with the gray body color occupied 55% and 45%, respectively.
The sex ratio of the females to the males in GRCC, GRCC1, and GGCC were approximately 1:1(P > 0.05), respectively (Table 5).
Fertility of GRCC
The mature testis of GRCC at the age of 6 months contained many lobules in which there were a lot of spermatozoa and some spermatogonia (Fig. 5a–c, e). The sperm consisted of a head and a tail (Fig. 5b). The head was surrounded by plasma membrane and had compact nuclear material (Fig. 5c). At the neck between the head and tail of the sperm, there were some mitochondria. In the tail, the representative construction of “9 + 2” canaliculus was observed (Fig. 5d). The spermatogonia contained the nucleus and the cytoplasm in which there a lot of ribosomes, endoplasmic reticulum, and mitochondria (Fig. 5e). Observed by scanning electron microscope, the spermatozoa of GRCC (Fig. 5f) showed the normal appearance with the head and the tail as that of RCC. The diameter of the head of the spermatozoa of GRCC was 1.9 μm, which was equal to that of RCC.
Structures of the mature testis and the appearance of the sperm of GRCC. a Histological section of the normal mature testis of GRCC, in which there were many lobules containing a lot of sperm (arrow); b ultrathin section of the mature testis of GRCC, in which in a lobule, there were a lot of mature sperm consisting of the head and tail (arrow); c the head of the mature sperm in the mature testis of GRCC was surrounded by plasma membrane and had compact nuclear material. At the neck between the head and tail of the sperm, there were some mitochondria (arrow); d in the tail of the sperm of GRCC, the representative construction of “9 + 2” canaliculus (arrow) was observed; e the spermatogonia (arrow) in the mature testis of GRCC contained the nucleus and the cytoplasm in which there a lot of ribosomes, endoplasmic reticulum, and mitochondria. f the appearance of the sperm (arrow) produced by the male of GRCC under the electron scanning microscope
The mature ovaries of 1-year GRCC consisted of many full-yolk ova and a few of oogonia.
The DNA Markers of Sox Genes
The PCR results based on the primers of HMG of Sox genes and the sequencing results showed that there were three DNA fragments (215, 617, and 1,958 bp) in RCC, two DNA fragments (215 and 616 bp) in GRCC and GRCC1, and three DNA fragments (215, 616, and 1,959 bp) in GGCC (Fig. 6). All the sequences of the PCR products have been submitted to GenBank, and their accession numbers were presented in Table 6. By comparing the sequences, we confirmed that 215 bp DNA fragment existed in RCC belonged to Sox 11, whereas 215 bp DNA in GRCC, GRCC1, and GGCC represented Sox 1 gene. The 616 bp DNA fragments existed in RCC, GRCC, GRCC1, and GGCC represented Sox 9a gene. The 1,958 bp and 1,959 bp DNA fragments in RCC and GGCC, respectively, were from Sox 4 gene.
Amplified DNA fragments resulted from the PCR based on the primers of HMG of Sox genes in RCC, GRCC, GRCC1, and GGCC. M DNA ladder markers with the increase of 200 bp. In lane 1, there were three DNA fragments (215, 617, and 1,958 bp) in RCC; in lane 2, there were two DNA fragments (215 and 616 bp) in GRCC; in lane 3, there were two DNA fragments (215 and 616 bp) in GRCC1; in lane 4, there were three DNA fragments (215, 616, and 1,959 bp) in GGCC
Table 7 indicated the percentage of nucleotide similarities of separate regions of the DNA fragments produced by PCR with the primers of HMG-box of Sox genes in GRCC, GRCC1, GGCC, and their maternal parent-RCC. In the sequences of the 215 bp DNA fragments in RCC, GRCC, GRCC1, and GGCC, 64.1% identity between RCC and GRCC, 64.6% identity between RCC and GRCC1, 62% identity between RCC and GGCC, 99.5% identity between GRCC and GRCC1, 97.6% identity between GRCC and GGCC, and 97.2% identity between GRCC1 and GGCC were found, indicating that the sequences of this DNA fragment in GRCC, GRCC1, and GGCC were lowly homologous to that of RCC (Table 7).
As for the sequences of the DNA fragments of 616 bp in RCC, GRCC, GRCC1, and GGCC, there existed the 98.7% similarity between RCC and GRCC, 98.8% similarity between RCC and GRCC1, 98.7% similarity between RCC and GRCC1, 99.8% similarity between GRCC and GRCC1, and 99.8% similarity between GGCC and GRCC1, indicating the sequences of this DNA fragment in GRCC, GRCC1, and GGCC were highly homologous to that of RCC (Table 7).
With regard to the sequences of the DNA fragment of 1958 bp in RCC, and 1,959 bp in GGCC, there were 99.8% similarity between RCC and GGCC, suggesting that the sequence in this DNA region of GGCC was very close to that of RCC (Table 7).
The Microsatellite DNA Markers
Among five pairs of primers (MFW1, MFW7, MFW15, MFW17, and MFW21) of microsatellite DNA, we found the primers of MFW7 were available to distinguish GRCC from RCC or BSB. Figure 7 showed the electrophotogram of microsatellite DNA patterns produced by the primer MFW7 in ten females of RCC, ten males of BSB, and five males and five females of GRCC. The results indicated that RCC and BSB could be detected by this pair of primers because they had quite different microsatellite DNA patterns. In GRCC, there existed the similar DNA fragments (arrows 4 and 5) with those in RCC, suggesting that GRCC inherited those DNA fragments from RCC. On the other hand, in GRCC, there also existed the similar DNA fragments (arrows 1 and 7) to those in BSB, suggesting that GRCC also inherited these DNA fragments from BSB. Interestingly, the new DNA fragments (arrows 2, 3, and 6) were only found in GRCC, neither in RCC nor in BSB, suggesting the DNA structure variances occurred in GRCC. The DNA fragments similar to those of BSB and the new DNA fragments were observed both in three (Fig. 7, nos. 11–12; no.15) out of five males and one (Fig. 7, no. 16) out of five females of GRCC, suggesting that these DNA fragments were not related to the sex determination.
Electrophotogram of microsatellite DNA patterns produced by the primer MFW7 in ten females of RCC, ten males of BSB, and five males and five females of GRCC. Lanes 1–10 represented ten females of RCC. Arrows 1 and 2 indicated the specific DNA bands only found in RCC, neither in GRCC nor in BSB. Lanes 11–20 represented five males (nos. 11–15) and five females (nos. 16–20) of GRCC. Arrows 1 and 7 indicated the DNA bands GRCC and BSB commonly had, but not found in RCC, suggesting that these DNA fragments came from the genome of BSB. Arrows 2, 3, and 6 indicated the specific DNA bands only found in GRCC, neither in RCC nor in BSB, suggesting the DNA structure variances occurred in GRCC. Lanes 21–30 represented ten males of BSB. Arrows 1–3 indicated the specific DNA bands only found in BSB, neither in RCC nor in GRCC. C represented the negative control. M represented the pBR322 DNA/Mspl Marker
Discussion
The distant crossing is an important and effective means to increase genetic variation in the hybrid progeny. With this method, it is possible to form the polyploid hybrids including the fertile tetraploid hybrids and sterile triploid hybrids by translating the whole genome from one species to another species. In addition, it is possible to form the bisexual diploid gynogenetic fish with the genetic variation in the form of the microchromosomes as the genetic leakage of the paternal fish, in which there is no treatment for inactivating the sperm and doubling the chromosome number as the artificial gynogenesis requires (Sun et al. 2007).
In the present study, the ploidy level of the gynogenetic fish was confirmed by counting chromosomal number (Fig. 3b–d), forming the karyotype (Fig. 3e), and FISH analysis (Fig. 4c). All the above results were in agreement to determine that the gynogenetic fish was diploid with 100 chromosomes and one to three microchromosomes. The microchromosomes are also called B chromosomes, which have been described in more than 1,300 species of plants and almost 500 species of animals (Camacho et al. 2000). The interspecific hybridization events were considered as one of the reasons for the origin of the microchromosomes in these natural species of plants and animals (Camacho et al. 2000). In the gynogenetic fish, P. formosa, the tiny microchromosomes were found to be derived from the males of closely related species (Poecilia mexicana, Poecilia latipinna, and Poecilia latipunctata; Lamatsch et al. 2004). However, it is not clear what the real male is. In this study, we not only found that there existed the tiny microchromosomes in GRCC but also provided the direct evidence that the tiny microchromosomes were derived from the males of BSB, supporting the opinion that the hybridization events can result in the formation of the microchromosomes.
The proposal that B chromosomes in one species could have originated from the A chromosomes of a closely related species was described in some cases (Camacho et al. 2000). For example, the spontaneous origin of B chromosomes was found in interspecific crosses between Coix aquaticus and Coix gigantean (Sapre and Deshpande 1989). This phenomenon was also found in the gynogenetic fish P. formosa. Laboratory crosses between individuals of P. formosa and males of a black strain, both lacking B chromosomes, generated some black-pigmented offspring, most likely the result of paternal pigmentation genes located on B chromosomes which appeared in the offspring because of incomplete elimination of paternal A chromosomes (Schartl et al. 1995). In the present study, it is obvious that the microchromosomes in GRCC and their offspring originated from the A chromosomes of BSB.
What is the reason for the formation of the bisexual fertile diploid gynogenetic fish? It is possible to contribute to the natural selection for better survival for the hybrids. In the catalog, the RCC and BSB belonged to different subfamilies. Their chromosomal number 100 (RCC) and 48(BSB) had so big difference that their adult diploid hybrids were lethal, and only the diploid embryos (2n = 74) survived (Fig. 3a). To overcome the chromosome and gene incompatibilities, the following three approaches occurred. First, the 74 (50 + 24) chromosomes as the diploid hybrids were spontaneously doubled to 148 ({50 + 24} ×2) chromosomes as the tetraploid hybrids (Liu et al. 2007b), in which RCC and BSB had its own homologue chromosomes by which the chromosome and gene incompatibilities were largely reduced. Second, the triploid hybrids with 124 (50 × 2 + 24) chromosomes was formed (Liu et al. 2007b), in which RCC had its own homologue chromosomes, and the chromosome and gene incompatibilities were reduced on some extent. Third, the 50 chromosomes in the egg of RCC were spontaneously doubled to 100 (50 × 2) chromosomes after its second polar body was retained. The diploid eggs bearing 100 chromosomes had the potential to develop into the living gynogenetic fish. And in some gynogenetic fish, the paternal genetic leakage occurred which formed one to three microchromosomes. The existence of the microchromosomes indicated the hybridization effect and showed that the formation of GRCC was not the strict gynogenesis. Our results showed that the diploid hybrids (2n = 74), diploid gynogenetic fish (2n = 100), triploid hybrids (3n = 124), and tetraploid hybrids (4n = 148) accounted for 7.5%, 14%, 15.5%, and 63%, respectively, in all, suggesting that with the increase of the DNA content, the survival frequency of the hybrids went up. It also suggested that the hybrids with higher ploidy level had stronger ability to overcome the chromosome and gene incompatibilities and were more available for survival. Although the frequency of the gynogenetic fish was lower (14%), there existed both the male and female individuals which guaranteed they were able to reproduce their progeny.
What is the mechanism of the formation of the male gynogenetic fish? In our previous study (Sun et al. 2007), we made the artificial gynogenesis of RCC in which the UV-treated sterile sperm of BBS was used to activate the eggs of RCC to develop following the cold shock (0–4°C) for 30 min for inhibiting the ejection of the second polar body so as to double the egg’s chromosomes. The artificial gynogenetic progeny was all-female, neither male being found nor microchromosome being observed. The main differences between the natural gynogenesis in the present study and the previous artificial gynogenesis are the UV treatment to the sperm of BSB and the cold shock to the eggs of RCC where the UV treatment makes the sperm of BSB genetically sterile. In the present study, the sperm of BSB, which was not treated by UV, was able to enter the eggs of RCC. So it was possible for the genome of the sperm of BSB to recombine with the genome of RCC, which could lead to genetic variances via recombination. The hybridization can boost genetic variance, allowing colonization of unexploited niches (Mallet 2007). However, due to the chromosome and gene incompatibilities, most of the haploid genome of BSB’ sperm was inactivated and finally degraded, and some DNA fragments formed the microchromosomes incorporating into the eggs. When a microchromosome containing the male-determining gene from the paternal fish remained in an egg of RCC, the egg would develop into the male gynogenetic fish after its chromosomes were spontaneously doubled. The fertility of the male GRCC was proved by the structure of the testes (Fig. 5a, b), the appearance of the sperm (Fig. 5f), and the formation of GRCC1 and GGCC. One to three microchromosomes were found not only in all the male diploid gynogenetic individuals but also in some female ones, suggesting that the microchromosome(s) in the female ones did not contain the male-determining gene from the paternal fish. On the other hand, one to three microchromosomes were found in GRCC1 and GGCC, indicating that their microchromosomes were inherited from GRCC.
In GRCC, there existed approximately 1:1 sex ratio of the females to males. It was concluded that a chromosome of BSB containing the male-determining gene entered, with 50% probability, into an egg of RCC, which finally remained in the form of microchromosome containing the male-determining gene in GRCC and led to 50% male. Similarly, a microchromosome containing the male-determining gene in GRCC was distributed into the next generation with 50% probability to form 50% male.
The genotypic variances were found in GRCC. Regarding FISH, we only found one strong and one weak hybridizing signals in GRCC, while there were two strong and one weak hybridizing signals in RCC (Fig. 4c), suggesting that there existed some DNA variances in 5S rDNA in GRCC. It was probably attributed to the recombination of the chromosomes between the BSB and RCC. Furthermore, based on the DNA fragment markers using the primers of HMG-box Sox genes, RCC presented three DNA bands, whereas GRCC only showed two DNA bands without the specific 1,958 bp DNA band representing Sox 4 gene RCC possessed, suggesting that the recombination of the chromosomes between BSB and RCC led to the deletion of Sox 4 gene in GRCC. On the other hand, the reappearance of Sox 4 gene in GGCC meant that the meiotic recombination of the chromosomes of GRCC led to this change. In the sequences of the 215 bp DNA fragments in RCC, GRCC, GRCC1, and GGCC, the identity of those between RCC and GRCC and GRCC’s progeny was quite lower (<65%) than that (>97%) between GRCC and GRCC’s offspring, also suggesting that there existed some DNA variances in this DNA fragment of GRCC.
The results of the microsatellite DNA markers in the present study indicated that in GRCC, not only the DNA fragments (Fig. 7, arrows 4 and 5) similar to those of RCC were found, but also the DNA fragments (Fig. 7, arrows 1 and 7) similar to those of BSB were observed, suggesting that GRCC contained the DNA fragments from BSB. Furthermore, in GRCC, the new DNA fragments (Fig. 7, arrows 2, 3, and 6) were not found either in RCC nor in BSB, thus suggesting the DNA structure variances occurred in GRCC.
In GRCC, not only the genotypic variances occurred, but also the phenotypic variances happened. For example, the differences (P > 0.01) were significant in the ratio of body width to body length between GRCC (0.52) and RCC (0.41), in the ratio of head width to head length between GRCC (0.92) and RCC (0.85), and in the dorsal fin number between GRCC (16–17) and RCC (18–20). The traits such as the wider body and wider head GRCC had were similar to those of BSB, not similar to those of RCC. As for the feeding habit, like that of BSB, GRCC and their progeny were herbivores. On the other hand, GRCC (F1) produced the diversified offspring (F2): GRCC1 and GGCC, whereas the RCC only generated the identical offspring RCC and does not generate any diversified progeny, indicating that GRCC was genetically different from RCC, and that the males of GRCC were genotypic males of gynogenesis. The presence of GGCC with the gray body color which was similar to body color of BSB meant that the DNA fragment of BSB bearing the gray-color-determining gene remained in GRCC in the form of microchromosome. So, we think it is the hybridization effect that makes GRCC change genetically and morphologically.
In the present study, the very important point is that both the fertile females and males of GRCC (F1) were found. Within 1 year, the males and females of GRCC reached maturity and generated the progeny (F2) after their mating. The fertility of the male gynogenetic fish was confirmed by the testis’s micro-and ultra-structure observation (Fig. 5a–c), the sperm shape observed by the electron scanning microscope (Fig. 5f), and the formation of the progeny (F2; Figs. 1 and 2). The sex ratio of the females to males in GRCC and their progeny (F2: GRCC1 and GGCC) was all close to 1:1 (P > 0.05), suggesting that GRCC, GRCC1, and GGCC had the potential to reproduce their progeny and form two new types of diploid fish population: GRCC1 and GGCC. The formation of GGCC with the gray body color suggested the distant hybridization of red crucian carp with related species with gray body color such as BSB could produce diploid gray crucian carp.
The formation of the bisexual GRCC and their progeny is significant in both evolutionary biology and fish genetic breeding. The diversity of the two kinds of progeny, GRCC1 and GGCC, suggest that they have the potential to become two new types of improved crucian carp with genetic variances, which is of great significance in evolutionary biology. In fish genetic breeding, GRCC and GRCC1 can be used as the new type of ornamental fish with red body color. GGCC can be used as the new type of aquaculture fish.
References
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. Phil Trans R Soc Lond B 355:163–178
Chen L, Li W, Liu SJ, Tao M, Long Y, Duan W, Zhang C, Xiao J, Qin QB, Luo KK, Liu JF, Liu Y (2009a) Novel genetic markers derived from the DNA fragments of sox genes. Mol Cell Probes 23:157–165
Chen SL, Tian YS, Yang JF, Shao CW, Ji XS, Zhai JM, Liao XL, Zhuang ZM, Su PZ, Xu JY, Sha ZX, Wu PF, Wang N (2009b) Artificial gynogenesis and sex determination in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol 11:243–251
Crooijmanns RPMA, Bierbooms VAF, Komen J, Poel VDJJ, Groenen MAM (1997) Microsatellite markers in common carp (Cyprinus carpio L). Anim Genet 28:129–134
Hubbs C, Drewry GE (1959) Occurrence and morphology of a phenotypic male of a gynogenetic fish. Science 129:1227–1229
Lamatsch DK, Nanda I, Schlupp I, Epplen JT, Schmid M, Schartl M (2004) Distribution and stability of supernumerary microchromosomes in natural populations of the Amazon molly, Poecilia formosa. Cytogenet Genome Res 106:189–194
Levan A, Fredga K, Sandburg A (1964) Nomenclature for centromeric positions on chromosomes. Hereditas 52:201–220
Li Y, Cai M, Wang Z, Guo W, Liu X, Wang X, Ning Y (2008) Microsatellite-centromere mapping in large yellow croaker (Pseudosciaena crocea) using gynogenetic diploid families. Mar Biotechnol 10:83–90
Liu SJ, Liu Y, Zhou GJ, Zhang XJ, Luo C, Feng H, He XX, Zhu GH, Yang H (2001) The formation of tetraploid stocks of red crucian carp × common carp hybrids as an effect of interspecific hybridization. Aquaculture 192:171–186
Liu JF, Liu SJ, Tao M, Li W, Liu Y (2007a) Isolation and expression analysis of testicular type Sox9b in allotetraploid fish. Mar Biotechnol 9:329–334
Liu SJ, Qin QB, Xiao J, Lu WT, Shen JM, Li W, Liu JF, Duan W, Zhang C, Tao M, Zhao RR, Yan JP, Liu Y (2007b) The formation of the polyploid hybrids from different subfamily fish crossings and its evolutionary significance. Genetics 176:1023–1034
Mallet J (2007) Hybrid speciation. Nature 446:279–283
Masaru M, Hideo F (1998) Characterization of repetitive DNA sequences carrying 5S rDNA of the triploid ginbuna (Japanese silver crucian carp, Carassius auratus langsdorfi). Genes Genet Syst 73:9–20
Nanda I, Schlupp I, Lamatsch DK, Lampert KP, Schmid M, Schartl M (2007) Stable inheritance of host species-derived microchromosomes in the gynogenetic fish Poecilia formosa. Genetics 177:917–926
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York, pp 463–468
Sapre AB, Deshpande DS (1989) Origin of B chromosomes in Coix L. through spontaneous interspecific hybridization. J Hered 78:191–196
Schartl M, Nanda I, Schlupp I, Wilde B, Epplen JT, Schmid M, Parzefall J (1995) Incorporation of subgenomic amounts of DNA as compensation for mutational load in a gynogenetic fish. Nature (London) 373:68–71
Sun YD, Zhang C, Liu SJ, Tao M, Zeng C, Liu Y (2006) Induction of gynogenesis in Japanese crucian carp (Carassius cuvieri). Acta Genetic Sinica 33:405–412
Sun YD, Tao M, Liu SJ, Zhang C, Duan W, Shen JM, Wang J, Zeng C, Long Y, Liu Y (2007) Induction of gynogenesis in red crucian carp using spermatozoa of blunt snout bream. Prog Nat Sci 17:163–167
Yu XJ (1989) China freshwater fisheries chromosome. Science Publishing House, Beijing, pp 44–75
Acknowledgments
This research was supported by the National Natural Science Fund for Distinguished Young Scholars (no. 30725028); State Key Basic Research and Development Program of China (No. 2007CB109200); and Specially-appointed Professor for Lotus Scholars Program of Hunan Province.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, S., Qin, Q., Wang, Y. et al. Evidence for the Formation of the Male Gynogenetic Fish. Mar Biotechnol 12, 160–172 (2010). https://doi.org/10.1007/s10126-009-9219-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-009-9219-9