Abstract
Fish species vary in their capacity to biosynthesize the n-3 long-chain polyunsaturated fatty acids (LC-PUFA), eicosapentaenoic (EPA) and docosahexaenoic (DHA) acids that are crucial to the health of higher vertebrates. The synthesis of LC-PUFA involves enzyme-mediated fatty acyl desaturation and elongation. Previously, a complementary DNA (cDNA) for an elongase, now termed elovl5a, had been cloned from Atlantic salmon. Here, we report on the cloning of two new elongase cDNAs: a second elovl5b elongase, corresponding to a 294-amino-acid (aa) protein, and an elovl2-like elongase, coding for a 287-aa protein, characterized for the first time in a nonmammalian vertebrate. Heterologous expression in yeast showed that the salmon Elovl5b elongated C18 and C20 PUFA, with low activity towards C22, while Elovl2 elongated C20 and C22 PUFA with lower activity towards C18 PUFA. All three transcripts showed predominant expression in the intestine and liver, followed by the brain. Elongase expression showed differential nutritional regulation. Levels of elovl5b and particularly of elovl2, but not of elovl5a, transcripts were significantly increased in liver of salmon fed vegetable oils (VO) compared to fish fed fish oil (FO). Intestinal expression showed a similar pattern. Phylogenetic comparisons indicate that, in contrast to salmon and zebra fish, Acanthopterygian fish species lack elovl2 which is consistent with their negligible ability to biosynthesize LC-PUFA and to adapt to VO dietary inclusion, compared to predominantly freshwater salmonids. Thus, the presence of elovl2 in salmon explains the ability of this species to biosynthesize LC-PUFA and may provide a biotechnological tool to produce enhanced levels of LC-PUFA, particularly DHA, in transgenic organisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In vertebrates, biosynthesis of long-chain polyunsaturated fatty acids (LC-PUFA) involves sequential desaturation and elongation of precursor essential PUFA, 18:2n-6 and 18:3n-3. Synthesis of arachidonic acid (ARA; 20:4n-6) is achieved by Δ6 desaturation of 18:2n-6 to produce 18:3n-6 that is elongated to 20:3n-6 followed by Δ5 desaturation (Cook 1996). Synthesis of eicosapentaenoic acid (EPA; 20:5n-3) from 18:3n-3 uses the same enzymes and pathway as for ARA, but docosahexaenoic acid (DHA; 22:6n-3) synthesis requires two further elongation steps, a second Δ6 desaturation and a chain-shortening step (Sprecher 2000). The extent to which any species can produce LC-PUFA varies and is dependent on their complement of fatty acyl desaturase and elongase enzymes. Freshwater fish and salmonids, including Atlantic salmon (Salmo salar), are capable of producing DHA from 18:3n-3 and so must express all the enzyme activities necessary for this biosynthetic pathway (Tocher 2003).
Interest in LC-PUFA synthesis in fish results from the fact that they are the primary source in the human food basket of the omega-3 or n-3 LC-PUFA that are crucial to the health of higher vertebrates. Aquaculture now supplies an increasing proportion of the fish for human consumption (Tidwell and Allan 2002; FAO 2006) and, until now, formulation of diets with fish oil (FO) has ensured that farmed fish are rich in n-3 LC-PUFA (Bell and Waagbø 2008). As global FO supplies are at their sustainable limit, further expansion of aquaculture requires suitable alternatives, with vegetable oils (VOs) being prime candidates. However, VOs are generally rich in C18 PUFA but lack the n-3 LC-PUFA abundant in FO (Sargent et al. 2002) and thus flesh of fish fed VO is characterized by increased levels of 18:2n-6 and 18:3n-3 and decreased levels of EPA and DHA, compromising their nutritional value to the human consumer (Izquierdo et al. 2003; Regost et al. 2003). Our hypothesis is that understanding the molecular basis of LC-PUFA biosynthesis and regulation in salmon will allow us to optimize the activity of the pathway to enable efficient and effective use of dietary VOs while maintaining the nutritional quality of the fish. Alimuddin et al. (2008) take this further by suggesting genetic engineering as a strategy to create fish capable of being reared on VO diets while still maintaining their health-promoting composition and, to demonstrate this, were able to increase EPA and DHA contents in transgenic zebra fish by overexpressing a masu salmon elovl5-like gene.
In mammals, several fatty acyl elongase genes termed ELOVL1 to ELOVL7, with differing fatty acid (FA) substrate specificities, have been described with ELOVL2 and ELOVL5 shown to participate in LC-PUFA biosynthesis (Leonard et al. 2000, 2002, 2004; Jakobsson et al. 2006). Mammalian ELOVL5 is predominantly involved in the elongation of C18 and C20 PUFA, whereas ELOVL2 has greatest activity in the elongation of C20 and C22 (Leonard et al. 2000, 2002, 2004). Previously, we cloned and characterized a complementary DNA (cDNA) for a fatty acyl elongase gene from Atlantic salmon (Hastings et al. 2005). Phylogenetic analysis grouped the salmon elongase cDNA into a cluster with greatest similarity to mammalian ELOVL5 (Leaver et al. 2008a). Heterologous expression in yeast showed that the product of the salmon elongase cDNA had the ability to lengthen C18 and C20 PUFA with only low activity towards C22 (Hastings et al. 2005). Therefore, the salmon elongase appeared to have a similar substrate specificity to mammalian ELOVL5. More recently, a search of the Atlantic salmon expressed sequence tag (EST) database (http://www.tigr.org) showed it contained a second fatty acyl elongase transcript that appeared related to mammalian ELOVL2 (Leaver et al. 2008a).
The aim of the present study was to isolate and characterize cDNAs for other fatty acyl elongases of Atlantic salmon, particularly for elovl2, which has not been previously reported in a nonmammalian vertebrate. Two new cDNAs identified as an elovl2-like elongase and a second elovl5-like fatty acyl elongase were cloned. Functional characterization of their encoded polypeptides in yeast, Saccharomyces cerevisiae, showed that the salmon Elovl2-like protein elongated C20 and C22 PUFA with only low activity towards C18 PUFA. The second elongase, named elovl5b, elongated C18 and C20 PUFA, with residual activity towards C22. The tissue distribution of the three elongase transcripts was determined, and their nutritional regulation in response to changes in the FA composition of the diet was analyzed in liver and intestine.
Materials and Methods
cDNA Cloning
The sequence of the previously cloned and characterized elovl5a cDNA (gb|AY170327|; Hastings et al. 2005) was used to query the GenBank dbEST (http://www.ncbi.nlm.nih.gov) and Atlantic salmon Gene Index (http://compbio.dfci.harvard.edu/tgi/) databases. This led to the identification of salmon EST sequences (gb|DW546112| and ti|TC91192|) showing similarity to the elovl5a cDNA. Primers specific to these sequences were designed and the fragments obtained by polymerase chain reaction (PCR) using GoTaq® Colorless Master Mix (Promega, Southampton, UK), following manufacturer’s instructions, were sequenced (CEQ-8800 Beckman Coulter Inc., Fullerton, U.A) and further extended by 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR (FirstChoice® RLM-RACE kit, Ambion, Applied Biosystems, Warrington, UK) to produce full-length cDNAs. These were deposited in the GenBank database under accession numbers gb|FJ237531| and gb|FJ237532| for the cDNA produces from the gb|DW546112| and ti|TC91192| ESTs, respectively.
Sequence and Phylogenetic Analysis
The deduced aa sequences of the two newly cloned elongase cDNAs were aligned with those of salmon (Ss) Elovl5a (NP_001117039) and of human (Hs) ELOVL5 (NP_068586) and ELOVL2 (NP_060240) using ClustalW2 or, to compare sequences two by two, the EMBOSS Pairwise Alignment Algorithms tool (http://www.ebi.ac.uk/Tools/emboss/align/) was used. The aa sequences of elovl genes from other fish species were derived by searching the ENSEMBL genome database using BLASTX and the protein sequences of human and salmon elovl5 and elovl2 as keys. Genomic sequences with similarity were extracted and compared with the protein sequence of salmon elovl2 and elovl5 using Wise2 (http://www.ebi.ac.uk/Tools/Wise2/index.html) to generate deduced Elovl polypeptides from Tetraodon nigroviridis, Takifugu rubripes, Danio rerio, Gasterosteus aculeatus, and Oryzias latipes. Deduced aa sequences of elongases from various species were aligned using ClustalX and sequence phylogenies were reconstructed using the neighbor-joining method (Saitou and Nei 1987). GenBank accession numbers for these sequences are: NP_956747 and NP_001035452 for D. rerio (Dr), AAV67803 for Oncorhynchus mykiss (Om), AAL69984 for Scophthalmus maximus (Sm), AAT81404 for Sparus aurata (Sa), AAO13174 for Oreochromis niloticus (On), AAT81406 for Gadus morhua (Gm), and AAT81405 for Clarias gariepinus (Cg). Human and Atlantic salmon sequences were the same used in the aa alignment. Confidence in the resulting phylogenetic tree branch topology was measured by bootstrapping through 1,000 iterations.
Functional Characterization
PCR fragments corresponding to the open reading frames (ORFs) of the putative elongases were amplified from salmon intestine cDNA using sense and antisense primers containing a digestion site (underlined)—Elo1BVF1 (CCCAAGCTTGAAATGGAGGCTTTTAATCATAAAC; HindIII) and Elo1BVR1 (CCGCTCGAGTCAGTCCACCCGCACTTT; XhoI), for gb|FJ237531|, and primers Elovl2VF2 (CCCGAGCTCAAGATGAACCATTTACAAAGTTTGG; SacI) and Elovl2VR1 (CCGCTCGAGCTACTTTCTCTTCTTGAAGCTG; XhoI) for gb|FJ237532|. PCR was performed using the high-fidelity PfuTurbo® DNA Polymerase (Stratagene, Agilent Technologies, Cheshire, UK), with an initial denaturing step at 95°C for 2 min, followed by 32 cycles of denaturation at 95°C for 30 s, annealing at 60°C (for primers Elo1BVF1/Elo1BVR1) or 62°C (for primers Elovl2VF2/ Elovl2VR1) for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min. The ORF of the salmon elovl5a cDNA was amplified using the primers described in Hastings et al. (2005). The DNA fragments were then digested with the corresponding restriction endonucleases (New England BioLabs, Herts, UK) and ligated into a similarly restricted pYES2 yeast expression vector (Invitrogen, Paisley, UK). Ligation products were used to transform Top10F’ Escherichia-coli-competent cells (Invitrogen) which were screened for the presence of recombinants. The purified plasmids (illustraTM GFX™ Micro Plasmid Prep Kit, GE Healthcare Life Sciences, Buckinghamshire, UK) containing the three elongase ORFs or the pYES vector alone were then used to transform S.-cerevisiae-competent cells (S.c. EasyComp Transformation Kit, Invitrogen). Transformation and selection of yeast with recombinant elongase–pYES2 plasmids, yeast culture, and FA analysis were performed as described in detail previously (Hastings et al. 2001, 2005; Agaba et al. 2004; Zheng et al. 2005a). Briefly, cultures of recombinant yeast were grown in S. cerevisiae minimal medium-uracil supplemented with one of the following FA substrates: stearidonic acid (18:4n-3), γ-linolenic acid (18:3n-6), EPA (20:5n-3), ARA (20:4n-6), docosapentaenoic acid (22:5n-3), or docosatetraenoic acid (22:4n-6). Docosapentaenoic and docosatetraenoic acids (>98–99% pure) were purchased from Cayman Chemical Co. (Ann Arbor, USA) and the remaining FA substrates (>99% pure) and chemicals used to prepare the S. cerevisiae minimal medium-uracil were from Sigma Chemical Co. Ltd. (Dorset, UK). FA were added to the yeast cultures at final concentrations of 2.5 (C18), 5.0 (C20) and 10.0 (C22) μM. After 2 days, yeasts were harvested and washed and lipid-extracted by homogenization in chloroform/methanol (2:1, v/v) containing 0.01% BHT as antioxidant. FA methyl esters were prepared, extracted, purified, and analyzed by gas chromatography (GC) in order to calculate the proportion of substrate FA converted to elongated FA product as \({\left[ {{{\text{product}}\;{\text{area}}} \mathord{\left/ {\vphantom {{{\text{product}}\;{\text{area}}} {{\left( {{\text{product}}\;{\text{area}} + {\text{substrate}}\;{\text{area}}} \right)} \times 100}}} \right. \kern-\nulldelimiterspace} {{\left( {{\text{product}}\;{\text{area}} + {\text{substrate}}\;{\text{area}}} \right)} \times 100}} \right]}\). The identity of the FA peaks was based on GC retention time and confirmed by GC–mass spectrometry as described previously (Hastings et al. 2001; Agaba et al. 2004).
Tissue Distribution and Nutritional Regulation
For the tissue expression profile, tissues (intestine, liver, white muscle, red muscle, kidney, spleen, heart, brain, gill, and adipose tissue) were dissected from three salmons, immediately frozen in liquid nitrogen, and stored at −70°C pending RNA extraction. The fish were ∼150 g post-smolts held in 7-m3 seawater tanks at ambient temperature, salinity, and photoperiod in the Marine Environment Research Laboratory, Machrihanish, Scotland, UK, and fed a commercial salmon feed based on fish meal and FO. The effects of diet on elongase expression were investigated in samples from salmon post-smolts fed four diets with the same basal composition but formulated with different oils, FO, rapeseed oil (RO), linseed oil (LO), or soybean oil (SO). Full descriptions of the diets and the trial have been reported previously (Leaver et al. 2008b). At the end of the trial, 0.5 g of liver and small intestine of five fish per dietary treatment were dissected and rapidly disrupted in 5 ml of TRI Reagent (Ambion, Applied Biosystems) using an Ultra-Turrax homogenizer (Fisher Scientific, Loughborough, UK) and immediately frozen in liquid nitrogen and stored at −70°C prior to RNA extraction.
Tissue RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted by organic solvent, according to manufacturer’s instructions (Ambion, Applied Biosystems), and RNA quality and quantity were assessed by electrophoresis (Bioanalyzer 2100, Agilent Technologies, Santa Clara, USA) and spectrophotometry (NanoDrop ND-1000, Thermo Scientific, Wilmington, USA), respectively. One microgram of total RNA per sample was reverse-transcribed into cDNA using a Verso™ cDNA kit (ABgene, Surrey, UK), following manufacturer’s instructions. Briefly, each 20-μl reaction contained 1 μg of total RNA, 300 ng of random hexamers and 125 ng of anchored oligo-dT, dNTP mix (500 μM each), 1× cDNA synthesis buffer, RT enhancer, and Verso enzyme mix. Following cDNA synthesis at 42°C for 1 h, reactions were stopped by heating at 95°C for 2 min and cDNA was diluted tenfold with water.
Expression of the three elongase transcripts was studied by quantitative real-time PCR (qPCR). The qPCR primers were designed in regions corresponding to the 3′ untranslated region of elovl5a and of the two new elongase cDNAs. In addition, amplification of four reference genes (elongation factor-1α (elf-1α), β-actin, an EST previously shown to be unresponsive to VO dietary manipulation, and 18S rRNA) was also performed, for normalization of the results. The unresponsive EST is an anonymous cDNA feature selected from a salmon cDNA microarray study and identified as a suitable reference gene on the basis of constant expression between different VO diets and time points (Taggart et al. 2008). Table 1 shows the sequence of the primers used, their specific annealing temperatures, the size of fragments produced, and the reference sequences used for primer design, using the Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). For tissue distribution, qPCR amplicons corresponding to the three putative elongases were cloned into the pBluescript® II KS phagemid vector (Stratagene), while that of 18S rRNA was cloned into the pCR2.1-TOPO vector (TOPO TA cloning kit, Invitrogen). These recombinant vectors were then restricted with SacI or XhoI and the linearized plasmid DNA containing the target sequence for each gene was quantified spectrophotometrically and serial-diluted to generate a standard curve of known copy numbers. For effects of diet, qPCR analysis used relative quantification with elf-1α, β-actin, and the unresponsive EST as reference genes, and amplification efficiency of the primer pairs was assessed by serial dilutions of cDNA pooled from the samples being quantified. qPCR amplifications were carried out in duplicate using a Quantica machine (Techne, Cambridge, UK) in a final volume of 20 μl containing 2 μl (for the nutritional regulation trial and 18S rRNA) or 5 μl (for tissue distribution) diluted (1/10) cDNA, 0.5 μM of each primer, and 10 μl AbsoluteTM QPCR SYBR® Green mix (ABgene). Amplifications were carried out with a systematic negative control (nontemplate control (NTC), containing no cDNA). The qPCR profiles contained an initial activation step at 95°C for 15 min, followed by 30 to 40 cycles: 15 s at 95°C, 15 s at the specific primer pair annealing Tm (Table 1), and 30 s at 72°C. After the amplification phase, a melt curve of 0.5°C increments from 75°C to 90°C was performed, enabling confirmation of the amplification of a single product in each reaction. The qPCR product sizes were checked by agarose gel electrophoresis and their identity was confirmed by sequencing. No primer–dimer formation occurred in the NTC.
Statistical Analysis
For tissue expression profiles, results were expressed as mean normalized values (±SD) corresponding to the ratio between the copy numbers of the putative elongase transcripts and the copy numbers of the reference gene, 18S rRNA. For the tissues with highest expression level, intestine and liver, a one-way analysis of variance followed by a Tukey honestly significant difference test, at a significance level of P < 0.05, was performed to compare the expression level of the three transcripts in both tissue samples, using Statistica 6 (StatSoft, Tulsa, USA). The effects of diet on elongase expression, expressed as the relative expression ratio of each gene in fish fed one of the VOs in relation to those fed FO (control) and normalized by three reference genes (elf-1α, β-actin, and the unresponsive EST), were analyzed for statistical significance using the relative expression software tool (REST-MCS©, version 2, http://www.gene-quantification.info/), which employs a pairwise fixed reallocation randomization test (10,000 randomizations) with efficiency correction (Pfaffl et al. 2004).
Results
Salmon Fatty Acyl Elongase cDNA Sequences and Phylogenetics
A 1,650-bp full-length cDNA sequence was obtained by 5′ and 3′ RACE PCR from gb|DW546112| and was deposited in the GenBank database under the accession number gb|FJ237531|. It contains an ORF of 885 bp encoding a putative protein of 294 aa, sharing 91% aa sequence identity with the previously described Elovl5-like salmon elongase (Elovl5a) and 93% identity in nucleotide sequence, in the ORF, to elovl5a (gb|AY170327|). This protein is 70% identical to several mammalian ELOVL5-like proteins (Rattus norvegicus, Mus musculus, and Homo sapiens) and 74–81% identical to other fish Elovl5-like proteins (D. rerio, G. morhua, O. niloticus, C. gariepinus, S. maximus, and S. aurata). This new transcript was thus named elovl5b.
For the sequence derived from ti|TC91192|, a full-length cDNA spanning 1,792 bp, with an 864-bp ORF encoding a putative 287-aa protein, was obtained and determined to have 79% aa sequence identity to the zebra fish Elovl2-like and 68–71% aa identity to mammalian (rat, mouse, and human) ELOVL2-like proteins. This sequence was deposited in the GenBank database under the accession number gb|FJ237532|.
Typically, these newly cloned Elovl5-like and Elovl2-like proteins both possess the diagnostic histidine box HXXHH motif (Fig. 1) conserved in all elongases and also characteristic of desaturase and hydrolase enzymes containing a di-iron-oxo cluster (Tvrdik et al. 2000; Jakobsson et al. 2006). In addition, they possess two lysine or arginine residuals at the carboxyl terminus, KXRXX in Elovl5 like and KKXX in Elovl2, which are proposed to function as endoplasmic reticulum (ER) retrieval signals (Jakobsson et al. 2006). By sequence comparison with a mouse ELOVL2 (Tvrdik et al. 2000), five putative transmembrane-spanning domains, containing hydrophobic aa stretches, can be predicted (Fig. 1). Noteworthy is the fact that, of the 17-aa residues found by Leonard et al. (2004) to be highly conserved across 22 members of the elongase family, one residue has been conservatively replaced in SsElovl5b (Leu245 replaced by Phe245).
ClustalW2 alignment of the deduced amino acid sequences of Atlantic salmon (S. salar; Ss) elongases, SsElovl5a (Hastings et al. 2005; NP_001117039) and the two newly cloned SsElovl5b and SsElovl2 (mRNA sequence deposited in the GenBank database under accession numbers gb|FJ237531| and gb|FJ237532|, respectively), together with human (H. sapiens; Hs) elongases, HsELOVL5 (NP_068586) and HsELOVL2 (NP_060240). Identical residues are shaded black and similar residues (based on the Gonnet matrix, using ClustalW2 default parameters) are shaded gray. Indicated are the conserved histidine box motif, characteristic of desaturases and hydrolase enzymes containing a di-iron-oxo cluster (underlined), five (I–V) putative membrane-spanning domains predicted by Tvrdik et al. (2000; underlined with dashed line), and, in bold, the lysine or arginine residuals proposed to function as ER retrieval signals (Jakobsson et al. 2006). An asterisk indicates the 17 aa residues found by Leonard et al. (2004) to be highly conserved across PUFA elongases
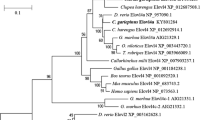
A phylogenetic tree was constructed on the basis of the aa sequence alignments between the putative two new salmon fatty acyl elongases and other elongases of fish and mammals (Fig. 2). The phylogenetic analysis showed that salmon Elovl5a and Elovl5b clustered with human ELOVL5 and several other fish proteins. In contrast, the putative salmon Elovl2 clustered separately from all other Elovl5 proteins and grouped with human ELOVL2 and an Elovl from zebra fish. BLASTX searches of the ENSEMBL genomes of T. nigroviridis, T. rubripes, G. aculeatus, and O. latipes demonstrated that these fish species do not contain elovl2 homologs.
Phylogenetic tree comparing putative amino acid sequences of Atlantic salmon, S. salar (Ss), Elovl2 and Elovl5b, cloned in this work, and that of Elol5a (NP_001117039), with those of other fatty acyl elongases: Human, H. sapiens (HsELOVL2 NP_060240 and HsELOVL5 NP_068586), zebra fish, D. rerio (Dr-NP_956747 and Dr-NP_001035452), trout, O. mykiss (Om-AAV67803), turbot, S. maximus (Sm-AAL69984), sea bream, S. aurata (Sa-AAT81404), tilapia, O. niloticus (On-AAO13174), cod, G. morhua (Gm-AAT81406), and catfish, C. gariepinus (Cg-AAT81405). The tree was constructed using the neighbor-joining method with ClustalX. Numbers represent the frequencies with which the tree topology presented was replicated after 1,000 bootstrap iterations. The scale refers to the horizontal branch length and corresponds to the amino acid substitution rate per site
Functional Characterization
The elongases were functionally characterized by determining the FA profiles of S. cerevisiae transformed with pYES2 vector alone or containing elovl5a, elovl5b, and elovl2 cDNA ORF inserts grown in the presence of potential FA substrates. The FA composition of the wild yeast insert is characterized by having essentially only 16:0, 16:1n-7, 18:0, and 18:1n-9 (see Hastings et al. 2001). Yeast transformed with vector containing no insert contains these fatty acids together with whichever exogenous FA was added, consistent with the well-established lack of PUFA elongase activity in S. cerevisiae (Agaba et al. 2004; Hastings et al. 2005). Heterologous expression of salmon elovl5a in yeast produced similar results to those previously observed (Hastings et al. 2005), with activity towards C18 and C20 substrates and only residual (1%) C22 to C24 activity (Table 2). The FA compositions of yeast transformed with the pYES2–elovl5b construct and grown in the presence of 18:4n-3, 20:5n-3, and 22:5n-3 are shown in Fig. 3a–c. The traces show the major endogenous FA (16:0, 16:1n-7, 18:0, and 18:1n-9, peaks 1–4) and additional peaks corresponding to the substrates and elongation products. Thus, exogenously added 18:4n-3 (peak 6) was elongated to 20:4n-3 (peak 7) and 22:4n-3 (peak 8; Fig. 3a); exogenously added 20:5n-3 (peak 9) was elongated to 22:5n-3 (peak 10; Fig. 3b), and only a trace of exogenously added 22:5n-3 (peak 10) was elongated to 24:5n-3 (peak 11; Fig. 3c). Other additional peaks including 18:1n-7 (peak 5), 20:1n-9, and 20:1n-7 indicated some capability of Elovl5b to elongate endogenous monounsaturated FAs, as previously observed with Elovl5a (Hastings et al. 2005). Elovl5b elongated 62% of 18:4n-3 and 71% of 18:3n-6, mainly to the C20 products (58% and 65% converted to 20:4n-3 and 20:3n-6, respectively), and 69% of 20:5n-3 and 48% of 20:4n-6, but only 1–2% of 22:5n-3 and 22:4n-6 were converted into 24:5n-3 and 24:4n-6 (Table 2). These data confirmed elovl5b as an ELOVL5-like elongase and its functional similarity to elovl5a.
Functional characterization of Atlantic salmon putative fatty acyl elongases, Elovl5b (a–c) and Elovl2 (d–f) in transgenic S. cerevisiae grown in the presence of n-3 FAs; 18:4n-3 (a, d); 20:5n-3 (b, e); and 22:5n-3 (c, f). Fatty acids were extracted from yeast transformed with pYES2 vector containing the ORF of the putative fatty acyl elongase cDNA as an insert. Peaks 1–4 represent the main endogenous FAs of S. cerevisiae, namely 16:0 (1), 16:1n-7 (2), 18:0 (3), and 18:1n-9 (4). Peak 5 corresponds to 18:1n-7 resultant from the elongation of the yeast endogenous16:1n-7 and the remaining main additional peaks (6–11) correspond to the exogenously added FAs and the products of their elongation—18:4n-3 (6), 20:4n-3 (7), 22:4n-3 (8), 20:5n-3 (9), 22:5n-3 (10), and 24:5n-3 (11). Other minor peaks are 18:2n-6, 20:1n-9, and 20:1n-7, the latter two resulting from the elongation of 18:1n-9 and 18:1n-7 (panels a–c). Vertical axis, FID response; horizontal axis, retention time
The FA compositions of yeast transformed with the pYES2–elovl2 construct and grown in the presence of FAs show a distinct pattern (Fig. 3d–f). Exogenously added 18:4n-3 (peak 6) was only poorly elongated to 20:4n-3 (peak 7; Fig. 3d), whereas exogenously added 20:5n-3 (peak 9) was elongated to 22:5n-3 (peak 10) and, especially, to 24:5n-3 (peak 11; Fig. 3e), and exogenously added 22:5n-3 (peak 10) was substantially elongated to 24:5n-3 (peak 11; Fig. 3f). Thus, only 5% and 12% of 18:4n-3 and 18:3n-6, respectively, were elongated, whereas 70% and 59% of 20:5n-3 and 20:4n-6, respectively, were elongated with 52% and 48% recovered as the C24 products (Table 2). Around 31% and 18% of exogenously added 22:5n-3 and 22:4n-6 were elongated to 24:5n-3 and 24:4n-6, respectively. Elongation of monounsaturated FAs was low, with conversion of 16:1n-7 to 18:1n-7 typically around 4%. No evidence for elongation of saturated FAs was observed with any of the salmon Elovls.
Tissue Distribution of Salmon Fatty Acyl Elongase cDNAs
All three elongase genes in salmon were expressed mostly in intestine (pyloric ceca) and liver, followed by brain (Fig. 4). The previously characterized elongase gene, elovl5a, showed a more widespread expression than the two new elongase transcripts being also found, albeit at a lower level in terms of absolute copy numbers, in gill, kidney, spleen, heart, adipose tissue, red muscle, and white muscle. In intestine, elovl5a and elovl2 appear to have a similarly high expression level, with elovl5b showing significantly lower expression. In liver, no significant differences were found, but copy numbers of elovl5b were slightly higher than those of the other two transcripts.
Tissue expression profile of elovl5a, elovl5b, and elovl2 in Atlantic salmon, determined by RT-qPCR. Absolute copy numbers were quantified for each transcript and were normalized by absolute levels of ribosomal 18s (values shown on top of each column and represented diagrammatically in logarithmic scale). I intestine, L liver, WM white muscle, RM red muscle, K kidney, SPL spleen, H heart, BR brain. Results are means (n = 3) ± SD. Different letters in intestine and liver columns indicate significant differences (P < 0.05) between transcripts in those tissues
Nutritional Regulation of Salmon Fatty Acyl Elongase Expression
The regulation of elongase genes, in response to dietary FA composition, was examined in liver and intestine of salmon that had been fed diets containing either FO rich in EPA and DHA or VOs rich in C18 FA, RO (18:1n-9), SO (18:2n-6), or LO (18:3n-3). Previously, transcriptomic analysis of samples from this VO-feeding experiment revealed changes in the transcript levels of genes in response to the FA composition of the diets but not of elongase (Leaver et al. 2008b). However, although the array contained elovl5a, both elovl5b and elovl2 were absent, and since elovl5a and the newly discovered elovl5b will cross-hybridize, it is not surprising that changes in expression were not detected by microarray analysis. However, LC-PUFA biosynthesis was elevated in these VO-fed salmon (Leaver et al. 2008b), and thus we examined the expression of all known salmon elovl genes in the present work through qPCR using gene-specific primers. Compared to the FO-fed group, there was a significant increase of elovl2 and elovl5b transcripts in the liver of VO-fed fish with normalized expression ratios for elovl2 of 2.2, 2.6, and 1.9 for treatments RO, SO, and LO, respectively, and a significant 1.7- and 1.5-fold increase of elovl5b transcript level in fish fed RO and SO diets, respectively (Fig. 5a). No significant differences were found between transcript levels for elovl5a in FO-fed fish and VO-fed fish or between the different VO dietary treatments for any of the genes. For intestine, elongase mRNA showed higher variability within dietary treatments than that observed in liver, and so no statistically significant differences were found in the relative expression of any of the elongase transcripts between fish fed the different dietary treatments (Fig. 5b). Nonetheless, the observed trends agreed with the results obtained in liver. Thus, the levels of elovl2 transcript in fish fed RO, SO, and LO diets were 3.0-, 4.2-, and 4.4-fold higher, respectively, compared to salmon fed FO. As observed in liver, the level of elovl5a transcripts in the intestine was unaffected in fish fed the different VO diets, when compared to the FO group, while RO- and SO-fed fish showed a 1.7- and 1.8-fold increase, respectively, in elovl5b transcript level compared to the FO treatment.
Nutritional regulation of elongase genes elovl5a, elovl5b, and elovl2, in Atlantic salmon fed diets containing fish oil (FO), rapeseed oil (RO), soybean oil (SO), or linseed oil (LO), in the liver (a) and intestine (b) tissues, determined by RT-qPCR. The results shown are the normalized expression ratio (reference genes: elf-1α, β-actin, and a flatliner EST) of the target transcripts in one of the VO treatments (RO, SO, or LO), in relation to the FO control treatment. Values are means (n = 5) with SE and asterisks represent significant differences (P < 0.05) between the column’s dietary treatment and the FO treatment, for the respective transcript (REST-MCS-2006)
Discussion
Very-long-chain fatty acyl elongases (ELOVL) are ER membrane-bound proteins responsible for the first regulatory step (condensation of activated FAs with malonyl-CoA) in the FA elongation pathway, elongating FA that are biosynthesized de novo or supplied by the diet. They belong to a gene family that consists of seven members in mice and humans, which differ in FA substrate specificity and have differing spatial and temporal expression patterns (Jakobsson et al. 2006). Of these seven ELOVL enzymes, ELOVL2 and ELOVL5 have been demonstrated to have a substrate preference for PUFA. In addition, ELOVL4 is critical for normal human and mouse retinal function and there is evidence to indicate that it may also be involved specifically in DHA biosynthesis, which is a major membrane component in these tissues (Zhang et al. 2001; Jakobsson et al. 2006). In mammals, ELOVL2 has greatest activity in the elongation of C20 and C22 but low or, in the case of human, no activity towards C18 PUFA (Leonard et al. 2002). In contrast, mammalian ELOVL5 is very active towards C18 PUFA but does not appear to have the capacity to elongate beyond C22 (Leonard et al. 2000; Inagaki et al. 2002). Other species for which a PUFA elongase gene has been reported and functionally characterized are the nematode Caenorhabditis elegans (Beaudoin et al. 2000) and fungus Mortierella alpina (Parker-Barnes et al. 2000), with these enzymes being predominantly active on C18 PUFA with virtually no activity towards C20, and the marine microalgae Pavlova, which has a unique specificity towards C20 with no C18 or C22 activity (Pereira et al. 2004).
Fish elovl cDNAs have been cloned and functionally characterized from a number of species: the freshwater species zebra fish, common carp, and tilapia; the salmonids, Atlantic salmon and rainbow trout; and the marine species cod, turbot, and sea bream (Agaba et al. 2004, 2005; Meyer et al. 2004; Hastings et al. 2005). More recently, a fatty acyl elongase from masu salmon has been cloned and overexpressed in zebra fish (Alimuddin et al. 2008). Phylogenetic analysis groups all these previously described elovl cDNAs into a cluster with greatest similarity to mammalian ELOVL5 (Meyer et al. 2004; Agaba et al. 2005; Alimuddin et al. 2008). All the fish elovl5 cDNAs tested lengthened monounsaturated FA and n-3 and n-6 PUFA with chain lengths from C18 to C22, with residual C22–C24 activity (Agaba et al. 2004, 2005; Hastings et al. 2005). A rainbow trout Elovl5 has also been described with C18 to C22 but no C22–C24 activity (Meyer et al. 2004). Thus, fish elovl5 appears to have wider PUFA specificity than its mammalian homolog, although activity towards C22 is minor, compared to C18 and C20 (Agaba et al. 2004, 2005; Hastings et al. 2005). No ELOVL2-like genes have been reported so far in a nonmammalian vertebrate. However, searches in the GenBank and Atlantic salmon EST databases, as well as in the zebra fish genome, demonstrated that a second PUFA elovl gene exists in Atlantic salmon and zebra fish and that this gene is clearly related to the mammalian ELOVL2 (Fig. 2). In addition, a second Atlantic salmon elovl5 gene, here named elovl5b to distinguish it from the previously cloned salmon gene (Hastings et al. 2005), now termed elovl5a, was identified. Preliminary work has revealed two distinct genomic sequences corresponding to these two transcripts, indicating that they are encoded by separate loci (unpublished results). Given the high degree of similarity of these two salmon elovl5 genes (91% and 93% identical at the protein and ORF nucleotide sequence level, respectively), we can speculate that they may result from a duplicated locus, as the result of the recent salmonid tetraploidization (Allendorf and Thorgaard 1984). This conclusion is supported by searches of ENSEMBL fish genomes, which contain only single copies of elovl5. Analysis of the deduced aa sequences of Elovl5b and Elovl2 showed that they possess characteristic features of microsomal membrane-bound enzymes, including a single histidine box redox center motif, a canonical ER retention signal (carboxyl-terminal dilysine/arginine targeting signal), and multiple transmembrane regions (Tvrdik et al. 2000; Jakobsson et al. 2006).
Functional characterization of the new elovl cDNAs by heterologous expression in yeast confirmed the identities predicted by sequence homology and phylogenetic analysis. Elovl5b had similar activity to the previously reported salmon Elovl5a (Hastings et al. 2005) and to that of other fish Elovls which clustered with mammalian ELOVL5 (Agaba et al. 2004, 2005). The only difference appears to be in terms of the relative conversion of n-3 and n-6 C18 (but not C20) FAs, as salmon Elovl5a had a preference for n-3 FA substrates, which was not apparent in Elovl5b. This preference seems variable as some fish Elovl5 proteins showed similar activities with n-3 and n-6 FAs, and cod Elovl5 was more active towards n-6 FAs (Agaba et al. 2005). The C18 and C20 PUFA specificity of both salmon Elovl5 proteins was generally comparable to that of mammalian ELOVL5 (Leonard et al. 2000; Inagaki et al. 2002) but a residual activity towards C22 was also measured. In addition, as observed with human and rat ELOVL5 homologs (Leonard et al. 2000; Inagaki et al. 2002), some capacity to elongate monounsaturated FAs was found for both salmon Elovl5 proteins, as in other fishes (Meyer et al. 2004; Agaba et al. 2005). More importantly, the other salmon cDNA described here is the first elovl2-like elongase to be cloned and functionally characterized in fish. In comparison to mouse ELOVL2 (Leonard et al. 2002), the salmon Elovl2 protein showed similar low activity towards C18 and high activity towards C20 and C22 but had a low capacity to convert monounsaturated FAs (around 4% conversion of 16:1n-7), which was not observed in mammalian ELOVL2 (Leonard et al. 2002). Notably, the major difference in comparison to fish Elovl5 and mammalian ELOVL5 proteins is the higher activity towards C22 LC-PUFA exhibited by Elovl2.
Other fish ESTs potentially corresponding to putative elovl2 transcript are those of zebra fish (gb|NM_001040362|), Pimephales promelas (gb|DT135746|; partial cDNA), and Ictalurus punctatus (gb|BM438219.1|; partial cDNA), all Ostariophysan freshwater species. Although the analysis of fish ELOVL genes is not yet exhaustive, searches in the puffer fish, stickleback, and medaka genomes have not indicated the presence of elovl2-like genes and thus it is possible to speculate that all fish of the order Acanthopterygii lack ELOVL2 homologs. Therefore, the characterized elovl5 cDNAs of sea bream, turbot, and cod would be the sole PUFA elovl gene in these species. Elovl5 proteins have a very limited capacity towards C22 and because biosynthesis of DHA in vertebrates requires elongation from C22 to a C24 PUFA intermediate, followed by a peroxisomal β-oxidation chain-shortening step (Buzzi et al. 1997; Sprecher 2000), fish which lack elovl2 would be restricted in their ability to produce this essential FA. In fact, where LC-PUFA biosynthesis has been directly measured in fish, it is clear that salmonids have substantially greater capacity than marine fish, such as sea bass, a member of the Acanthopterygii (Mourente et al. 2005). Previously, the ability of both salmon and zebra fish to produce LC-PUFA endogenously has been shown to be due to the presence of both Δ5 and Δ6 desaturase genes (Hastings et al. 2005; Zheng et al. 2005a), which contrasts with the single Δ6 gene so far discovered in Acanthopterygii (Tocher et al. 2006). Here, we further show that the varying competences of different fish to biosynthesize LC-PUFA, particularly DHA, might not only depend on their genome complement of desaturase genes but also of elongases. The phylogenetic distribution of the elovl2 gene suggests that it must have been lost from the evolutionary line leading to the Acanthopterygii but retained in Ostariophysi (e.g., zebra fish) and Salmoniformes. It is tempting to speculate that, since both Ostariophysi and Salmonid fish spend all or at least a substantial period of time in freshwater, the retention of elovl2 and the diversification of desaturase genes are adaptations to the relative deficiency of preformed LC-PUFA in their freshwater habitats compared to marine ecosystems, which are fueled by LC-PUFA-producing phytoplankton (Sargent et al. 2002; Brett and Muller-Navarra 1997). Although also speculative, it can be observed that the vast majority of Acanthopterygian species inhabit marine environments. Those that inhabit freshwater tend to live in productive lacustrine or large riverine systems. Few, if any, exist in the nutrient-poor streams and lakes that salmonids inhabit.
The tissue distribution profile of salmon elovl5a had been reported previously and agreed broadly with that found here (Zheng et al. 2005a). The tissue distribution of elovl5b largely resembled that of elovl5a, apart from significantly lower transcript levels in intestine, slightly higher in liver, and lower in brain. In general, elovl5a, elovl5b, and elovl2 are expressed predominately in intestine and liver. Since these tissues are the major sites of lipid synthesis and distribution, this expression profile is consistent with a role in FA biosynthesis. In the remaining tissues tested, elovl5a was the most highly expressed. Rat and human ELOVL5 was expressed in most tissues examined (Inagaki et al. 2002; Leonard et al. 2000; Wang et al. 2005) and ELOVL2 transcripts, as in salmon, appeared to have a more restricted distribution in mammals (Leonard et al. 2002; Tvrdik et al. 2000; Wang et al. 2005). Salmon brain exhibited the third highest expression of the tissues analyzed but expression levels of elovl2 were very low, over one order of magnitude lower than elovl5b and even less than elovl5a. This might mean that DHA, which comprises a great proportion of the brain’s cellular membranes, either has an exogenous source or that another gene, such as a homolog to the mammalian ELOVL4, for instance, is involved in DHA biosynthesis in the brain. This hypothesis should be investigated in the future.
This work has clearly shown that salmon elovl5b and elovl2, but not elovl5a, transcripts are increased in response to inclusion of VO in the diet. However, previous studies examining the effect of FO replacement by VO in the diets of Atlantic salmon on elovl5a expression have yielded inconsistent results (Zheng et al. 2004, 2005a, b). In mammals, ELOVL5 and ELOVL2 expression and regulation have been described. Compared to Elovl5, levels of Elovl2 mRNA were low in rat liver (Wang et al. 2005), while in salmon these genes are expressed at similar levels. If these transcript levels are reflected in protein concentrations, then some differences in the relative importance and contribution of elovl2 and elovl5 to LC-PUFA synthesis in salmon and rats might be expected. In regard to nutritional regulation, Inagaki et al. (2002) reported a lack of dietary regulation, by cycles of fasting–refeeding, while Wang et al. (2005) observed that overnight starvation and feeding FO-enriched diets decreased Elovl5 mRNA levels in rat liver. No diet-induced changes were noted in Elovl2 expression (Wang et al. 2005). Thus, Elovl5 rather than Elovl2 appeared to play the major role (along with Δ5 and Δ6 desaturases) in response to changes in dietary lipid composition in rat (Wang et al. 2005), while elovl2 may have a more prominent role in salmon. This might not be surprising, considering the differences in dietary regimes of mammals and carnivorous fish, particularly concerning the relative supply of C20 and C22 FAs.
Hepatic Elovl2 and Elovl5 are both regulated by sterol regulatory element binding protein (SREBP) transcription factors in mouse (Horton et al. 2003) and SREBP activation is increased by low cholesterol and decreased by increased PUFA (Espenshade 2006), suggesting potential mechanisms for the observed changes in expression of these genes in salmon fed VO diets which are low in both PUFA and cholesterol (Leaver et al. 2008b). These increases in elovl2 and elovl5 are important in salmon aquaculture because they enable physiological adaptation to VO diets which are deficient in FA greater than 18 carbons and enable LC-PUFA biosynthesis. Moreover, the observed difference in the nutritional regulation of elovl5a and elovl5b genes will provide an interesting opportunity to advance our knowledge on the regulation and transcriptional control of gene expression, by comparison of the presumably very similar upstream regulatory regions of these genes that have resulted from a recent gene duplication event.
In conclusion, two elovl genes, homologs of mammalian ELOVL2 and ELOVL5, have been characterized in Atlantic salmon, which together with the previously characterized salmon Δ5 and Δ6 fatty acyl desaturases, explains the ability of these fish to biosynthesize LC-PUFA, including DHA, more efficiently than Acanthopterygian fish species which seem to lack an elovl2 gene. This and the ability of salmon to regulate these genes in response to dietary quality might to some extent also explain the success of salmonids in colonizing nutrient-poor freshwater habitats and to better tolerate VO-based aquaculture diets than cultured Acanthopterygians. The results obtained are not only of high relevance in advancing our understanding of the molecular basis of LC-PUFA biosynthesis and regulation in fish but also have biotechnological significance. The use of transgenic techniques to enhance EPA and DHA biosynthesis is likely to become routine in various organisms of commercial importance (Meyer et al. 2004). A previously cloned and characterized zebra fish desaturase (Hastings et al. 2001) has already been utilized to produce transgenic Arabidopsis capable of producing EPA and DHA at low levels (Robert et al. 2005). The identification of salmon elovl2, a fatty acyl elongase with high activity in the penultimate steps of LC-PUFA biosynthesis, might provide a route for increasing EPA and DHA production in transgenic organisms. Indeed, interest is also likely to become directed towards optimizing LC-PUFA biosynthesis pathways through transgenic techniques in farmed fish to enable efficient and effective use of dietary VO while maintaining the nutritional quality of the fish as a primary source in the human food basket of the omega-3 or n-3 LC-PUFA. Alimuddin et al. (2008) have already increased EPA and DHA biosynthesis in zebra fish by overexpressing an elovl5-like gene from masu salmon. Given the differences in substrate specificity of elovl5 and elovl2, reported here for the first time in fish, and the necessity of C22 to C24 PUFA intermediate elongation for DHA biosynthesis (Sprecher 2000), the potential benefits of overexpressing these two genes in parallel can easily be recognized.
Abbreviations
- aa:
-
amino acid
- ARA:
-
arachidonic acid (20:4n-6)
- DHA:
-
docosahexaenoic acid (22:6n-3)
- EPA:
-
eicosapentaenoic acid (20:5n-3)
- ER:
-
endoplasmic reticulum
- FA:
-
fatty acid
- FO:
-
fish oil
- LC-PUFA:
-
Long chain polyunsaturated fatty acids (carbon chain length ≥ C20 with ≥3 double bonds)
- LO:
-
linseed oil
- ORF:
-
open reading frame
- PUFA:
-
polyunsaturated fatty acids
- qPCR:
-
quantitative (real-time) polymerase chain reaction
- RACE:
-
rapid amplification of cDNA ends
- RO:
-
rapeseed oil
- SO:
-
soybean oil
- UTR:
-
untranslated region
- VO:
-
vegetable oil
References
Agaba M, Tocher DR, Dickson C, Dick JR, Teale AJ (2004) Zebra fish cDNA encoding multifunctional fatty acid elongase involved in production of eicosapentaenoic (20:5n-3) and docosahexaenoic (22:6n-3) acids. Mar Biotechnol 6:251–261
Agaba MK, Tocher DR, Dickson CA, Zheng X, Dick JR, Teale AJ (2005) Cloning and functional characterisation of polyunsaturated fatty acid elongases from marine and freshwater teleost fish. Comp Biochem Physiol 142B:342–352
Alimuddin Kiron V, Satoh S, Takeuchi T, Yoshizaki G (2008) Cloning and over-expression of a masu salmon (Oncorhynchus masou) fatty acid elongase-like gene in zebra fish. Aquaculture 282:13–18
Allendorf FW, Thorgaard GH (1984) Tetraploidy and the evolution of salmonid fishes. In: Turner BJ (ed) Evolutionary genetics of salmonid fishes. Plenum, New York, pp 1–53
Beaudoin F, Michaelson LV, Lewis MJ, Shewry PR, Sayanova O, Napier JA (2000) Production of C20 polyunsaturated fatty acids (PUFAs) by pathway engineering: identification of a PUFA elongase component from Caenorhabditis elegans. Biochem Soc Trans 28:661–663
Bell JG, Waagbø R (2008) Safe and nutritious aquaculture produce: benefits and risks of alternative sustainable aquafeeds. In: Holmer M, Black KD, Duarte CM, Marba N, Karakassis I (eds) Aquaculture in the ecosystem. Springer, Heidelberg, pp 185–225
Brett MT, Muller-Navarra DC (1997) The role of highly unsaturated fatty acids in aquatic food web processes. Freshwater Biol 38:483–499
Buzzi M, Henderson RJ, Sargent JR (1997) Biosynthesis of docosahexaenoic acid in trout hepatocytes proceeds via 24-carbon intermediates. Comp Biochem Physiol 116:263–267
Cook HW (1996) Fatty acid desaturation and chain elongation in eukaryotes. In: Vance DE, Vance JE (eds) Biochemistry of lipids, lipoproteins and membranes. Elsevier, Amsterdam, pp 129–152
Espenshade PJ (2006) SREBPs: sterol-regulated transcription factors. J Cell Sci 119:973–976
Food and Agricultural Organisation, FAO (2006) State of world aquaculture 2006. FAO Fisheries Technical paper No. 500, FAO, Rome
Hastings N, Agaba M, Tocher DR, Leaver MJ, Dick JR, Sargent JR, Teale AJ (2001) A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc Natl Acad Sci U S A 98:14304–14309
Hastings N, Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ (2005) Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar). Mar Biotechnol 6:463–474
Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A 100:12027–12032
Inagaki K, Aki T, Fukuda Y, Kawamoto S, Shigeta S, Ono K, Suzuki O (2002) Identification and expression of a rat fatty acid elongase involved the biosynthesis of C18 fatty acids. Biosci Biotechnol Biochem 66:613–621
Izquierdo M, Obach A, Arantzamendi L, Montero D, Robaina L, Rosenlund G (2003) Dietary lipid sources for sea bream and sea bass: growth performance, tissue composition and flesh quality. Aquacult Nutr 9:397–407
Jakobsson A, Westerberg R, Jacobsson A (2006) Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res 45:237–249
Leaver MJ, Bautista JM, Björnsson BT, Jönsson E, Krey G, Tocher DR, Torstensen BE (2008a) Towards fish lipid nutrigenomics: current state and prospects for fin-fish aquaculture. In: Sundell K, Power D (ed) State-of-the-art scientific and/or technologic reviews on the use of molecular methods and functional genomics in aquaculture. Rev Fish Sci 16(Supplement 1):73–94
Leaver MJ, Villeneuve LAN, Obach A, Jensen L, Bron JE, Tocher DR, Taggart JB (2008b) Functional genomics reveals increases in cholesterol biosynthetic genes and highly unsaturated fatty acid biosynthesis after dietary substitution of fish oil with vegetable oils in Atlantic salmon (Salmo salar). BMC Genomics 9:299. doi:10.1186/1471-2164-9-299
Leonard AE, Bobik EG, Dorado J, Kroeger PE, Chuang L-T, Thurmond JM, Parker-Barnes JM, Das T, Huang Y-S, Murkerji P (2000) Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem J 350:765–770
Leonard AE, Kelder B, Bobik EG, Chuang L-T, Lewis CJ, Kopchick JJ, Murkerji P, Huang Y-S (2002) Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37:733–740
Leonard AE, Pereira SL, Sprecher H, Huang Y-S (2004) Elongation of long-chain fatty acids. Prog Lipid Res 43:36–54
Meyer A, Kirsch H, Domergue F, Abbadi A, Sperling P, Bauer J, Cirpus P, Zank TK, Moreau H, Roscoe TJ, Zähringer U, Heinz E (2004) Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J Lipid Res 45:1899–1909
Mourente G, Dick JR, Bell JG, Tocher DR (2005) Effect of partial substitution of dietary fish oil by vegetable oils on desaturation and oxidation of [1-14C] 18:3n-3 and [1-14C]20:5n-3 in hepatocytes and enterocytes of European sea bass (Dicentrarchus labrax L.). Aquaculture 248:173–186
Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung L-T, Huang Y-S, Mukerji P (2000) Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc Natl Acad Sci U S A 97:8284–8289
Pereira SL, Leonard AE, Huang Y-S, Chuang L-T, Mukerji P (2004) Identification of two novel microalgal enzymes involved in the conversion of the ω3-fatty acid, eicosapentaenoic acid, into docosahexaenoic acid. Biochem J 384:357–366
Pfaffl MW, Horgan GW, Dempfle L (2004) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi:10.1093/nar/30.9.e36
Regost C, Arzel J, Robin J, Rosenlund G, Kaushik SJ (2003) Total replacement of fish oil by soybean or linseed oil with a return to fish oil in turbot (Psetta maxima). 1. Growth performance, flesh fatty acid profile, and lipid metabolism. Aquaculture 217:465–482
Robert SS, Singh SP, Zhou X-R, Petrie JR, Blackburn SI, Mansour PM, Nichols PD, Liu Q, Green AG (2005) Metabolic engineering of Arabidopsis to produce nutritionally important DHA in seed oil. Functional Plant Biol 32:473–479
Saitou N, Nei M (1987) The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sargent JR, Tocher DR, Bell JG (2002) The lipids. In: Halver JE, Hardy RW (eds) Fish nutrition. 3rd edn. Academic, San Diego, pp 181–257
Sprecher H (2000) Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta 1486:219–231
Taggart JB, Bron JE, Martin SAM, Seear PJ, Høyheim B, Talbot R, Carmichael SN, Villeneuve LAN, Sweeney GE, Houlihan DF, Secombes CJ, Tocher DR, Teale AJ (2008) A description of the origins, designs and performance of the TRAITS-SGP Atlantic salmon Salmo salar L. cDNA microarray. J Fish Biol 72:2071–2094
Tidwell JH, Allan GL (2002) Fish as food: aquaculture’s contribution. World Aquac 33:44–48
Tocher DR (2003) Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fisheries Sci 11:107–184
Tocher DR, Zheng X, Schlechtriem C, Hastings N, Dick JD, Teale AJ (2006) Highly unsaturated fatty acid synthesis in marine fish; cloning, functional characterization and nutritional regulation of fatty acid Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids 41:1003–1016
Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A (2000) Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol 149:707–717
Wang Y, Botolin D, Christian B, Busik J, Xu J, Jump DB (2005) Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases. J Lipid Res 46:706–715
Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ, Kakuk LE, Lagali PS, Wong PW, MacDonald IM, Sieving PA, Figueroa DJ, Austin CP, Gould RJ, Ayyagari R, Petrukhin K (2001) A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Gen 27:89–93
Zheng X, Tocher DR, Dickson CA, Bell JG, Teale AJ (2004) Effects of diets containing vegetable oil on expression of genes involved in highly unsaturated fatty acid biosynthesis in liver of Atlantic salmon (Salmo salar). Aquaculture 236:467–483
Zheng X, Tocher DR, Dickson CA, Dick JR, Bell JG, Teale AJ (2005a) Highly unsaturated fatty acid synthesis in vertebrates: new insights with the cloning and characterisation of a Δ6 desaturase of Atlantic salmon. Lipids 40:13–24
Zheng X, Torstensen BE, Tocher DR, Dick JR, Henderson RJ, Bell JG (2005b) Environmental and dietary influences on highly unsaturated fatty acid biosynthesis and expression of fatty acyl desaturases and elongase genes in liver of Atlantic salmon (Salmo salar). Biochim Biophys Acta 1734:13–24
Acknowledgements
This work and XZ were supported by the Biotechnology and Biological Sciences Research Council Responsive Mode Grant, BB/C51237X/1 (Transcriptional control of polyunsaturated fatty acid synthesis in fish). SM was supported by “Fundação para a Ciência e a Tecnologia”, Portugal (grant SFRH/BPD/34247/2006) and OM by the postdoctoral research program of the Fundación Española para la Ciencia y la Tecnología (Ministerio de Educación y Ciencia).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morais, S., Monroig, O., Zheng, X. et al. Highly Unsaturated Fatty Acid Synthesis in Atlantic Salmon: Characterization of ELOVL5- and ELOVL2-like Elongases. Mar Biotechnol 11, 627–639 (2009). https://doi.org/10.1007/s10126-009-9179-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-009-9179-0