Abstract
In this work, we propose to the Raman spectroscopy as a new technique for the detection of the type 2 diabetes using blood serum samples. The serum samples were obtained from 15 patients who were clinically diagnosed with type 2 diabetes mellitus and 20 healthy volunteers. The average spectra showed equally intense peaks as, 695 cm−1, the doublet of tyrosine at 828 and 853 cm−1, phenylalanine at 1002 and 1028 cm−1, the phospholipid shoulder at 1300–1345 cm−1, and proteins (amide I) at 1654 cm−1. The major differences were found at 661 and 1404 cm−1 (glutathione), 714 (polysaccharides), 605 (Phe), 545 cm−1 (tryptophan), and the shoulder of amide III at 1230–1282 cm−1, where seem to disappear in the diabetes spectrum. On the contrary, the region that is more highlighted due to that diabetes peaks are clearly more intense was 897–955 cm−1. Principal component analysis and linear discriminate analysis were employed for developing discrimination method. The first three principal components provided a classification of the samples from healthy and diabetes patients with high sensitivity and specificity. In addition, when the first principal component was plotted as a function of the Raman shift, it revealed these shifts accounted for the greatest differences between control and diabetes samples, which coincided with the shifts of spectral differences shown by mean spectra. Our results demonstrated that serum sample Raman spectroscopy promises to become a non-invasive support tool of the currently applied techniques for type 2 diabetes detection, decreasing the false-positive cases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes is a chronic and progressive disease that occurs either when the pancreas does not produce enough insulin, a hormone that regulates blood sugar or glucose, or when the body cannot effectively use the insulin it produces, increasing blood glucose levels. The World Health Organization (WHO) recognizes three forms of diabetes mellitus: type 1, type 2, and gestational diabetes (occurs during pregnancy), each with different causes and with different incidence [1, 2]. Type 1 diabetes is characterized by deficient insulin production in the body, due to the destruction of the β cells of the Islets of Langerhans of the pancreas, and people who survive with this diabetes requires insulin injections to regulate the amount of glucose in their blood. In type 2 diabetes, the body cannot properly use the insulin it produces.
In 2014, according to WHO, worldwide about 422 million adults were living with diabetes compared to 108 million in 1980 [1]. The global prevalence of diabetes has nearly doubled since 1980, rising from 4.7 to 8.5% in the adult population. Diabetes caused 1.5 million deaths in 2012, being the percentage of deaths, attributable to high blood glucose or diabetes that occurs prior to age 70, higher in low- and middle-income countries than in high-income countries. Diabetes and its complications bring about substantial economic loss to local health systems and national economies, so that it has become a priority for action by all countries.
The patient’s starting point to live well with diabetes is early diagnosis, which is based on continuous measurements of fasting plasma glucose concentration greater than or equal to 126 mg/dL (7.0 mmol/L). At present, the chemical test reagent strip read by a glucometer is the technique used to measure blood glucose concentrations [3]. Nevertheless, patients will require periodic specialist evaluations, treatment by complications, or sophisticated laboratory tests to distinguish between type 1 and type 2 diabetes. Therefore, there is considerable interest in developing both rapid, less invasive, and objective methods for the diagnosis and detection of diabetes and biochemical models that allow understanding the molecular changes that give rise to the disease.
In this sense, as an alternative to the conventional methods for the study of diabetes, the spectroscopic techniques have been explored. Thereby, fluorescence spectroscopy was used for discriminating between glycated and unglycated albumin diabetes markers [4]. The Fourier-transform infrared spectroscopy provided information of this important glycated analyte when incubating albumin with glucose [5, 6]. Rohleder et al. investigated serum samples of diabetes patients using mid-infrared spectroscopy [7]. However in the infrared technique, water, present in large proportions in the biological samples, shows a very complex spectrum because it is very sensitive to the vibrations and rotations of the water molecule [8]. In contrast, a spectroscopic technique not affected by the presence of water molecules, due to taking advantage of the low absorption coefficient of water in the near-infrared spectral region, is the Raman spectroscopy [9]. Therefore, Raman spectroscopy is the best technique for studying body fluids containing considerable amounts of water; furthermore that in it, no sample preparation is required. Dingari et al. used serum sample Raman spectroscopy and multivariate classification techniques to differentiate glycated albumin from the unglycated variant with 100% accuracy [10]. Raman spectroscopy detected minute amounts of glucose in diluted urine, with a 92% accuracy to classify abnormal and normal urine samples according to their glucose concentrations [11].
In addition to the invaluable results in the characterization of samples related to diabetes, Raman spectroscopy and multivariate analyses have impacted the detection of other degenerative diseases such as cervical and breast cancer using serum samples with high sensitivity and specificity [12, 13]. Furthermore, monitoring of leukemia chemotherapy treatment using Raman spectroscopy was possible through the exclusive use of blood serum samples [14]. Cancer results are markedly improved when the Raman technique is implemented with gold and silver nanoparticles, a technique known as surface enhanced Raman spectroscopy (SERS) technique [15, 16].
In this context, we propose in this article a new method of type 2 diabetes detection based on serum sample Raman spectroscopy and multivariate analyses. Firstly, Raman spectra are classified using principal component analysis (PCA) and as we know a priori how many groups there are and which samples correspond to each group, subsequently we applied linear discriminate analysis (LDA) as a technique acting in a supervised manner. In PCA, the positions of the main peaks, which are responsible for the separation of the spectral data for the formation of groups or clusters of data (data classification), can be calculated by plotting the first principal components as a function of the wavenumber [20]. These positions allow finding the most relevant biochemical differences between the samples of the control and diabetes patients. To the best of our knowledge, this is the first report of preliminary results evaluating the usefulness of Raman spectroscopy in the diagnosis of type 2 diabetes using serum samples.
Methodology
Samples
Fresh serum samples were obtained from 15 patients who were clinically diagnosed with type 2 diabetes mellitus and 20 healthy volunteer controls. All patients were from the western central region of Mexico and had similar ethnic and socioeconomic backgrounds. The age for the diabetes patients was between 40 and 65 years. The diabetes and control serum samples were obtained through the Human Ethical Committees of Mexican hospitals. Written consent was obtained from the subjects and the study was conducted according to the Declaration of Helsinki.
Blood samples were obtained between 7:00 and 9:00 A. M. and were centrifuged to get the serum. All spectra were obtained on the same day. The samples were frozen at − 189 ∘C in a liquid nitrogen dewar before Raman spectroscopy analysis was performed. To ensure statistically sound sampling, five spectra from different regions of each serum sample were collected. A total of 182 spectra were collected with 102 spectra from 20 control patients, 80 spectra from diabetes patients.
Raman spectroscopy
All spectra were collected at a Jobin-Yvon LabRAM HR800 Raman Spectrometer with a laser wavelength of 830 nm. A drop of serum was placed onto an aluminum substrate, which was examined by an Olympus microscope coupled to the Raman system and several points were chosen for Raman measurement with an exposure of 20 to 40 s. The laser beam was focused on the surface of the sample with a 100 × objective. The radius of the beam was 1.0 μ m and the laser power irradiation over the samples was 17 mW. The Raman system was calibrated with a silicon semiconductor using the Raman peak at 520 cm−1. All spectra were taken collected in the region from 400 to 1800 cm−1, with a resolution of 0.6 cm−1.
Data analysis
The average of Raman spectra taken per patients in control and diabetes groups was 5.1 and 5.3, respectively. Raw spectra were processed by carrying baseline correction, smoothing, and normalization to remove noise, sample florescence, and shot noise from cosmic rays, and then analyzed using PCA [17]. After the initial processing, the mean spectrum of each group was calculated. The mean spectra were analyzed to obtain general biochemical information for each data group [18, 19].
In order to apply the PCA, the matrix of spectra, built with the 182 Raman spectra, was used to calculate the covariance matrix, which in turn allowed decomposing large spectral data into small number of independent variations known as principal components (PCs) and whose contributions are known as scores. Score of components is one of the widely used parameters for classification. In the plot of the first principal component, it is expected to observe two large groups of points or spectra, groups of spectra of the control and diabetes patients. To guarantee maximum separability and therefore a more precise definition of the groups obtained in the PCA, the ratio of between-groups variance to the within-groups is maximized (LDA method).
By analyzing the PCA result through the LDA application, it is possible to count true-positive (TP) and false-negative (FN) cases for calculating the sensitivity of the method of diabetes diagnosis. Similarly, counting the true-negative (TN) and false-positive (FP) cases, the specificity of the method is calculated. Sensitivity and specificity are used to judge diagnostic ability.
Finally, for a cross-check to the comparative analysis of the average control and diabetes spectra, the loading vectors are plotted as a function of the wavenumber by determining the positions of relevant differences between groups of patients.
PCA, LDA, and all the algorithms for data analysis were implemented in MatLab commercial software.
Results
One hundred and eighty-two spectra were collected from 20 control and 15 type 2 diabetes serum samples.
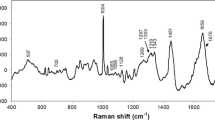
Figure 1 shows the comparison of mean Raman spectra from control and type 2 diabetes samples observing significant differences. The control serum spectrum showed the presence of higher amounts of carotenoids indicated by peaks at 1002, 1160, and 1523 cm−1 and intense peaks associated with protein components at 853, 938, 1002, 1300 to 1345, 1447, 1550, 1620, and 1654 cm−1. The major differences between diabetes and control spectra were observed with strong increases in the intensity of the bands 446 (glutathione), 897 (C-O-C str), 938 (protein components), 955 (CH2 rock) cm−1, and minor increases at 1523 (β carotene), 1556 (Trp), 1587 (vibrational modes of backbone and amino acid residues of proteins), 1603 (Tyr and Phe), 1620 (Tyr, Trp and C = C str) cm−1 in diabetes spectrum. Furthermore, strong decreases occurred at 622 (Phe) and 642 (C-S stretching vibration of Tyr) cm−1, and minor decreases at 509, 1063 (Phe), 1083 (phospholipid), 1126 (protein and lipids), 1174 (Trp and Phe), 1208 (Trp), and 1447 (lipids) cm−1 in diabetes spectrum. On the other hand, unchanged peak at 695, 828 and 853 (Tyr), 1002 and 1028 (Phe), 1160 (β carotene), the phospholipid shoulder at 1300–1345 and 1654 (Amide I) cm−1 between control and diabetes spectra. Finally, the bands at 545 (Trp), 605 (Phe), 661 and 1404 (glutathione), 714 (polysaccharides), 742 (phospholipid), and 1230–1282 (amide III) cm−1 seemed to disappear in the diabetes spectrum.
Table 1 shows the main bands observed in the control and type 2 diabetes spectra and the corresponding assignment of biomolecules. The last column of this table specifies the average spectra where the molecules are observed and that allowed discriminating the spectrum of diabetes samples from the spectrum of the control samples. The following section shows the results obtained by the PCA method and that determines those molecules that allowed discriminating the diabetes samples from the control samples but based on the analysis of all 182 spectra and not only of the two mean spectra of the diabetes and control samples.
PCA results
Once the spectra of the serum samples are collected, the next task is to analyze them as a whole, which can result in an extremely complex task when dealing with spectra of biological samples. For this reason, multivariate statistical tools, such as PCA, artificial neural network (ANN), and hierarchial cluster analysis (HCA), are often used to analyze this type of spectrum.
In this study, spectra of all two classes of samples, namely, control and diabetes, were pooled and analyzed by PCA to obtain discrimination among the classes. Plot of the first three principal components obtained in the region 400–1800 cm−1 is shown in Fig. 2.
We knew a priori how many groups there were and which samples corresponded to each group. In Fig. 2, the blue dots corresponded to the 102 spectra of the control patients and the red dots corresponded to the 80 spectra of the diabetes patients. We applied the multivariate technique, LDA, to our PCA result as a technique acting in a supervised manner. LDA identified the two most natural clusters in the total of 182 spectra by defining in the space of the principal component’s two well-localized zones (see Fig. 2). These zones watched with the two large group of patients reported by the diabetes specialist from the medical center, one zone containing the blue points (control cluster) and another zone containing the red points (diabetes cluster).
The number TN was determined by the number of blue points (control spectra) fully contained in the zone defined by our LDA as controls (left zone in Fig. 2), i.e., TN = 99. In the same way, the number TP was determined by the number of red dots (diabetes spectra) completely contained in the right zone in Fig. 2 defined by our LDA as diabetes, i.e., TP = 79. Only 1 of 80 spectra from diabetes serum samples was misclassified as control (FP = 1) and 3 of 102 spectra from control serum samples were misclassified as diabetes (FN = 3). Therefore, diagnostic specificity and sensitivity were 99 and 96%, respectively.
PCA was applied to discriminate between the Raman spectra of serum from control and diabetes patients using cross-validation. In cross-validation, the data was randomly split into two sets, a training set and a test set. In this approach, one sample (testing data) at a time was left out and PCA was applied after data reduction. Ten components for smoothing without baseline correction spectra and twelve components for smoothing with baseline correction spectra were considered for this analysis. In both cases, we were able to observe the two large groups of spectra as we obtained in Fig. 2. The sensitivity and specificity for data with smoothing and baseline correction and for smoothed data without baseline correction were 96 and 99%, respectively.
To bring out the differences in spectral profiles more clearly, the positions of relevant difference spectra were computed by plotting the first principal component as a function of the wavenumber [20]. According to custom, the principal differences between groups were represented by peaks with higher intensity. Nevertheless, several of these high peaks could be representing natural biochemical differences among only control patients. In order to know these natural differences, we plotted the first principal component versus the wavenumber between the 102 control spectra. Figure 3 shows control-control plots with the position of the most relevant differences between the control patients and control-diabetes plots with the position of the most relevant differences between the control and diabetes patients using only the first principal component, PC1. By discarding the most intense peaks matching between the control-control and control-diabetes plots (as 509, 558, 592, 642, 747, 1002, 1310, 1565, 1654 cm−1, among others), we obtained real biochemical differences among the control and diabetes serum samples. These differences appeared at 446, 451 (glutathione), 497 (Trp), 520, 566, 605 (Phe), 622 (Phe), 760 (Trp), 828 (glutathione), 853 (Phe), 1014 (glutathione), 1053, 1126 (protein), 1149, 1160 (β carotene), 1174 (Tyr), 1208 (Trp), 1274 (amide III), 1404 (glutathione), 1523 (β carotene), 1588 (protein, Tyr), 1602 (Tyr), 1620 (Tyr, Trp), and 1703 (glutathione) cm−1. Other differences can be determined using the principal components PC2 and PC3. As can be observed in Fig. 3, it could be an alternate method for viewing the differences in intensity observed by the loading vectors of PC1.
Discussion
Our preliminary study suggests that Raman spectroscopy has the potential to be a non-invasive diagnostic tool of type 2 diabetes with high sensitivity and specificity. It requires no sample preparation and provides objective, specific, and fast results.
The Raman bands assigned to glutathione (661 and 1404 cm−1), tryptophan (545 cm−1), phenylalanine (605 cm−1), polysaccharides (714 cm−1), phospholipid (742 cm−1), and protein amide III (1230 − 1282 cm−1) seemed to disappear in the diabetes spectrum whereas that the 897–955 cm−1 region is highlighted since several peaks of diabetes are clearly more intense suggesting that these features may play a role in the type 2 diabetes detection.
Raman spectroscopy and multivariate analyses were able to differentiate between diabetes and control samples with 96% sensitivity and 99% specificity. The first three principal components allowed observing two well-defined clusters of spectra corresponding to the types of samples used in our study. Therefore, a new patient can be diagnosed quickly and with high probability, by simply observing the cluster where his spectra fall. If the spectra fall into the diabetes cluster, then the patient will be diagnosed as a diabetes patient. In addition, these methods offer an alternative way to the analysis of the average spectra to determine the main biochemical differences between the control and diabetes samples. Plot, PC1 vs Raman shift, showed that the main peaks that generated the formation of the well-defined clusters of control and diabetes spectra were in agreement with the peaks obtained by directly observing the average spectra. By analyzing the plots corresponding to the components PC2 and PC3, other coincident and non-coincident peaks between the control and diabetes spectra can be determined.
Our study suggests, in analogy to the article reported by Haka et al. [21] where it presents the first demonstration of the in vivo collection of Raman spectra of breast tissue, that it is possible to design an effective method of diagnosing type 2 diabetes based on the collection of a large number of Raman spectra of blood serum samples from different patients officially diagnosed by medical institutions. This collection of spectra will allow monitoring diabetes patients under insulin treatment observing the biochemical changes during the treatment and the improvement of the patient when the group of spectra of the diabetes patients approaches the group of spectra of the control patients [14]. Our article is the starting point for the creation of this large library of molecular fingerprints of diabetes.
In addition, these results support the development of a rapid and low-cost method to discriminate between type 1 and type 2 diabetes samples using Raman spectroscopy in early stages of the disease.
Conclusion
Our preliminary results demonstrated that Raman spectroscopy and principal component analysis can be used to discriminate between serum samples from diabetic and healthy patients with high sensitivity and specificity. The study revealed that the main molecular differences between diabetes and control serum samples were glutathione, tryptophane, tyrosine, β carotene, and amide III. These biomolecules could play an important role in the early type 2 diabetes detection. The results of this exploratory study demonstrated that serum sample Raman spectroscopy promises to become a non-invasive support tool of the current techniques applied for type 2 diabetes detection by reducing the number of diagnostic tests and opens a great expectation for designing a Raman method that allows discriminating between type 1 and type 2 diabetes patients.
References
World Health Organization (2016) Global report on diabetes. ISBN 978 92 4 156525 7 (NLM classification: WK 810)
Cefalu WT (2016) Diabetes care. J Clin Appl Res Educ 39(Suppl 1):S13–S22. https://doi.org/10.4067/S0034-98872012001100015. ISBN 0149-5992
Hoenes J, Mueller P, Surridge N (2008) The technology behind glucose meters: test strips. Diab Technol Ther 10:S10–S26
Sakurai T, Takahashi H, Tsuchiya S (1984) New fluorescence of nonenzymatically glucosylated human serum albumin. FEBS Lett 176:27–31
Rondeau P, Navarra G, Cacciabaudo F, Leone M, Bourdon E et al (2010) Thermal aggregation of glycated bovine serum albumin. Biochim Biophys Acta 1804:789–798
Chesne S, Rondeau P, Armenta S, Bourdon E (2006) Effects of oxidative modifications induced by the glycation of bovine serum albumin on its structure and on cultured adipose cells. Biochimie 88:1467–1477
Rohleder D, Kocherscheidt G, Gerber K, Kiefer W, Köhler Möcks J, Petrich WW (2005) Comparison of mid-infrared and Raman spectroscopy in the quantitative analysis of serum. J Biomed Opt 10(3):031108
Max J-J, Chapados C (2009) Isotope effects in liquid water by infrared spectroscopy. III. H 2 O and D 2 O spectra from 6000 to 0 cm−1. J Chem Phys 131:184505
Carey DM (1998) Measurement of the Raman spectrum of liquid water. J Chem Phys 108:2669. https://doi.org/10.1063/1.475659
Dingari NC, Horowitz GL, Kang JW, Dasari RR, Barman I (2012) Raman spectroscopy provides a powerful diagnostic tool for accurate determination of albumin glycation. PLoS ONE 7(2):e32406. https://doi.org/10.1371/journal.pone.0032406
Park CS, Kim KK, Choi JM, Park KS (2007) Classification of glucose concentration in diluted urine using the low-resolution Raman spectroscopy and kernel optimization methods. Physiol Meas 28(5):583–593
González-Solís JL, Martínez-Espinosa JC, Torres-González LA, Jave-Suárez LF, Aguilar-Lemarroy AC, Palomares-Anda P (2014) Cervical cancer detection based on serum samples Raman spectroscopy. Lasers Med Sci 29:979–985
Pichardo-Molina JL, Frausto-Reyes C, Barbosa-García O, Huerta-Franco R, González-Trujillo JL, Ramírez-Alvarado CA, Gutiérrez-Juárez G, Medina-Gutiérrez C (2006) Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med Sci 10103:432–438
González-Solís JL, Martínez-Espinosa JC, Salgado-Román JM, Palomares-Anda P (2014) Monitoring of chemotherapy leukemia treatment using Raman spectroscopy and principal component analysis. Lasers Med Sci 29:1241–1249
Sánchez-Rojo SA, Martínez-Zérega BE, Velázquez-Pedroza EF, Martínez-Espinosa J C, Torres-González LA, Aguilar-Lemarroy A, Jave-Suárez LF, Palomares-Anda P, González-Solís JL (2016) Cervical cancer detection based on serum sample surface enhanced raman spectroscopy. Rev Mex Fis 62:213–218
Vargas-Obieta E, Martínez-Espinosa JC, Martínez-Zérega BE, Jave-Suárez LF, Aguilar-Lemarroy A, González-Solís JL (2016) Breast cancer detection based on serum sample surface enhanced Raman spectroscopy. Lasers Med Sci 31:1317–1324
Boelens HF, Eiler PH, Hankemeier T (2005) Sing constrains improve the detection of differences between complex spectral data sets: LC-IR as an example. Anal Chem 77(24):7998–8007
Stone N, Kendall C, Smith J et al (2004) Raman spectroscopy for identification of epithelial cancers. Faraday Discuss 126:141–157
De Gelder J, De Gussem K, Vandenabeele P, Moens L (2007) Reference database of Raman spectra of biological molecules. J Raman Spectrosc 38:1133–1147
Nogueira VG, Silveira L (2005) Raman spectroscopy study of atherosclerosis in human carotid artery. J Biomed Opt 10:031117–1–031117-7
Haka AS, Volynskaya Z, Gardecki J et al (2006) In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res 66:3317–3322
Funding
The authors wish to thank Research Network of CONACYT, Soft Condensed Matter, for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
González-Solís, J.L., Villafan-Bernal, J.R., Martínez-Zérega, B.E. et al. Type 2 diabetes detection based on serum sample Raman spectroscopy. Lasers Med Sci 33, 1791–1797 (2018). https://doi.org/10.1007/s10103-018-2543-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-018-2543-4