Abstract
Antimicrobial resistance is a major public-health concern. We evaluate chlorhexidine role in selection of resistant Pseudomonas aeruginosa mutants and their antibiotic cross-resistance. Mutation frequency and mutation rate after short-term exposure to sub-inhibitory concentrations of chlorhexidine were compared to those after spontaneous chlorhexidine-exposure, in P. aeruginosa PAO1 strain. Chlorhexidine-resistant mutants were generated, either by serial passage in increasing chlorhexidine concentrations or by single exposure to lethal chlorhexidine concentration. The generated mutants were tested for cross-resistance to different antibiotics, by determination of minimum inhibitory concentrations (MIC). The accompanied phenotypic changes in membrane permeability, outer membrane proteins (OMP), and efflux function were evaluated. The effect of exposure to chlorhexidine on MexAB-OprM, MexEF-oprN, and MexXY efflux pumps expression was investigated. No significant change was recorded between the mutation frequencies and mutation rates after short-term exposure to sub-inhibitory concentrations of chlorhexidine and after spontaneous chlorhexidine-exposure, in P. aeruginosa PAO1 strain. Twelve stable mutants, with ≥ eight-fold increase in chlorhexidine MIC, were generated. Several mutants showed increase in the MIC of colistin, cefepime, ceftazidime, meropenem, ciprofloxacin, and amikacin; seven mutants expressed meropenem cross-resistance. This was accompanied by decreased outer membrane permeability and changes in OMP. Using efflux pump inhibitor, chlorhexidine resistance was reverted in most isolates. Exposure to sub-inhibitory concentration of chlorhexidine induced the expression of MexXY efflux pump. Some resistant mutants had overexpressed MexXY efflux pump. Chlorhexidine can select P. aeruginosa strains with antibiotic cross-resistance. This necessitates implementing special protocols for chlorhexidine use and re-evaluation of its benefit versus risk in personal-care products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance is undermining our ability to treat the ever-increasing range of infections. According to the center for disease control and prevention (CDC), resistant bacteria cause more than 2.8 million infections with 35,000 deaths per year, in the USA [1]. Antibiotic resistance approximately kills 700,000 people each year worldwide, and some experts predict that a continued rise in resistance would lead to 10 million people dying yearly, by 2050 [2].
Many studies addressed the role of irrational use of antibiotics as the main determinant of the spread of microbial resistance but fewer focused on the possible effect of the use of non-antibiotic antimicrobial agents.
Non-antibiotic antimicrobials are widely used as antiseptics and disinfectants as well as a component of commercial soaps and detergents. Their use can select resistant mutants with cross-antibiotic resistance; for example, the exposure of Pseudomonas aeruginosa to triclosan has led to the selection of multidrug-resistant (MDR) strains [3]. In addition, some clinically important antiseptics such as benzalkonium chloride and chlorhexidine gluconate—besides being a substrate of some efflux systems—induce the expression of some efflux pumps as MexCD-OprJ [4, 5]. In 2016, The U.S. Food and Drug Administration (FDA) suspended the use of certain non-antibiotic antimicrobial agents (e.g., triclosan) in medicated soaps, based on their ability to trigger cross-antibiotic resistance with no known benefit [6].
Chlorhexidine (CHX) is one of the most widely used biocides among antiseptics and disinfectants. It is also incorporated into different products as a preservative [7]. It was reported that the excessive use of CHX, in the management of patients with long-term bladder catheters, resulted in urinary tract infections by CHX-resistant Gram-negative bacteria (i.e., Pseudomonas aeruginosa); these strains were also MDR [8]. Similarly, Stein and colleagues reported reduced susceptibility to chlorhexidine in all carbapenem-resistant Klebsiella pneumoniae isolates [9]. Recently, Wand and his colleagues reported that exposure to CHX may be associated with mutations in the two-component regulator, phoPQ, of K. pneumoniae. These mutants had stable reduction in the minimum inhibitory concentration (MIC) of the last-resort antibiotic; colistin [10].
P. aeruginosa is a member of the “ESKAPE” group of microbes and is considered one of the most common nosocomial organisms with high mortality rates especially in critically ill and immunocompromised patients. It is an opportunistic human pathogen characterized by an intrinsic resistance to multiple antimicrobial agents, leaving only few treatment options and representing one of the greatest therapeutic challenges [11]. Carbapenem-resistant P. aeruginosa was classified as critical in the World Health Organization list of priority pathogens for drug development [12], while MDR P. aeruginosa was classified as a serious threat by the center for disease control and prevention [1].
Here, we test the effect of CHX in selection of P. aeruginosa with cross-resistance to different antibiotics and some associated phenotypic and molecular changes, such as the change in membrane permeability, the change in outer membrane proteins, efflux-mediated resistance, and the change in gene expression of MexXY efflux pump. Also, the ability of CHX to induce overexpression of different efflux pumps was evaluated.
Methods
Bacterial strain and growth condition
P. aeruginosa PAO1 (P. aeruginosa ATCC 47085) strain was kindly provided by Dr. Ahmed Sherif Attia [13]. Overnight cultures were grown in Muller-Hinton (MH) broth at 37 °C unless otherwise stated.
Determination of CHX MIC
CHX was used as hydrochloride salt (Cat. number 220557; Merk, Germany). The MIC of CHX was determined by broth microdilution method, according to the clinical and laboratory standards institute guidelines [14]. CHX stock solution was prepared as 0.5 g/L. Twofold serial dilutions were made volumetrically in 50 µL MH broth (Oxoid, UK) to obtain a final dilution range of 0.05 to 100 µg/mL after the addition of inoculum suspension. Overnight culture in MH broth was diluted to reach an optical density equivalent to that of 0.5 McFarland. The adjusted inoculum was then diluted 1:150 so that after inoculation each well contained 5 × 104 CFU in 50 µL of the diluted culture. The MIC was defined as the lowest concentration of drug that inhibited the visible growth of the organism after 24 h of incubation at 37 °C.
Growth pattern of P. aeruginosa PAO1 in presence of CHX
An overnight culture of P. aeruginosa PAO1 strain, in MH broth, was diluted 1:100 in 50 mL MH broth, containing either 0.25 or 0.5 the MIC of CHX (0.781 or 0.39 µg/mL, respectively). Culture in MH broth without CHX was tested simultaneously. All cultures were incubated at 37 °C with shaking at 180 rpm. The effect of CHX addition in mid-log phase (after 5 h of inoculation; t = 5 h) was also tested, by adding the same CHX concentrations (0.25 and 0.5 of the MIC) to the P. aeruginosa PAO1 strain cultures in MH broth after 5 h of incubation. Samples were withdrawn every hour for the first 10 h then after 24 h post-inoculation, and their optical density (OD) was measured at 600 nm. The growth curve was constructed by plotting the optical density of the culture versus time. The growth rate was calculated for the cultures grown either in absence of CHX or with CHX added at the start of experiment using the method described by Tsuchiya and colleagues [15].
Determination of the spontaneous mutation frequency
P. aeruginosa PAO1 was grown overnight to reach a viable cell number of about 109 CFU/mL. This culture was diluted to 106 CFU/mL (1:1000) with fresh MH broth. A 50 µL of the diluted broth was inoculated into each of 13 independent tubes containing 450 µL fresh MH broth and allowed to grow for 3 h, at 37 °C with shaking at 180 rpm, to obtain parallel independent cultures. Spontaneous resistant mutants were obtained by plating the whole culture (500 µL) onto MH agar plates (ten plates) containing 50 µg/mL CHX, as a selective concentration (32 times the MIC, maximum solubility).
The total number of cells per culture was determined by plating the appropriate dilutions of three cultures onto non-selective medium (MH agar plates). Colonies on both selective and non-selective plates were counted after incubation for 48 h at 37 °C. The mutation frequency (the resistance index) was expressed as the mean number of resistant cells divided by the total number of viable cells per culture [16]. The experiment was done as three independent biological replicates each with 13 parallel cultures (three of which were used for viable count determination and the remaining ten cultures were plated on CHX-containing plates).
Determination of the mutation frequency after short-term exposure to sub-inhibitory concentrations of CHX
P. aeruginosa PAO1 was grown overnight and diluted to 106 CFU/mL as described previously, then 50 µL of the diluted broth culture was inoculated into three sets of 13 independent 450 µL fresh MH broth tubes. Each set of the MH broth tubes contained CHX at a final concentration of either 0.125, 0.25, or 0.5 of MICs (0.2, 0.4, or 0.8 µg/mL, respectively). The cultures were incubated for 30 min at 37 °C with shaking at 180 rpm, to obtain parallel independent cultures.
The number of resistant mutants and the total number of cells as well as the mutation frequency (the resistance index) were determined as described previously.
Estimation of mutation rates after spontaneous and short-term exposure to CHX
Trial to estimate possible variations in mutation rates after spontaneous and short-term exposure to different concentrations of CHX, using the data generated during spontaneous mutation frequency and mutation frequency after short-term exposure to CHX, was done using the fluctuation assay. The mutation rates were calculated using the p0 method for determination of m, where m is the most likely number of mutations per culture. The mutation rate was then calculated to be equal to m/Nt, where Nt is the total number of cells per culture [17].
Generation of CHX-resistant mutants
CHX-resistant mutants were generated by serial passage in increasing concentrations of CHX. Briefly, different concentrations of CHX were prepared in 10 mL MH broth by serial dilution (0.2 to 12.5 µg/mL). They were inoculated by 10 µL of an overnight culture of P. aeruginosa PAO1 strain and incubated at 37 °C for 48 h.
The highest CHX concentration that showed positive growth was used to inoculate another set of MH broth with increasing CHX concentrations as mentioned above (0.2 to 12.5 µg/mL) and the same procedure was repeated until the tested culture was able to grow in a higher CHX concentration than its MIC, for three successive passages. This culture was tested for its CHX MIC, by broth microdilution method as described previously, and considered resistant phenotype from passage 1 (P1); P1 was stored in glycerol stock for further testing.
P1 was then isolated on MH agar plates and one colony was used to inoculate another set of increasing CHX concentrations as described before. The previous procedure was repeated until we were able to obtain a strain that can grow at 32 times the MIC of CHX (50 µg/mL).
In addition, ten colonies were picked randomly from different plates (mutation frequency plates), after exposure to CHX either spontaneously or after short-term exposure to sub-inhibitory concentrations of CHX; they were stored in glycerol for further testing.
Determination of the phenotypic stability of the obtained resistant phenotypes
The stability of resistant phenotypes, generated by single or continuous exposure to CHX, was examined. One isolated colony from each culture was subjected to serial passage in MH broth, without CHX, daily for ten consecutive passages. This was followed by determination of the MIC values by broth microdilution method, using the culture from the tenth passage as an inoculum. Cultures that maintained an elevated MIC of more than or equal to eightfold of the MIC of parent strain (≥ 12.5 µg/mL) were considered stable resistant mutants. Glycerol stocks were prepared from the obtained resistant mutants. Prior to any experiment, the test strain was retrieved by isolation on MH agar and its MIC was retested as described previously to confirm maintenance of the same level of CHX resistance.
Confirmation of the purity of the mutant strain
The purity of the mutant strains was tested phenotypically, by API 20 NE (analytical profile index, BioMerieux, USA) following the manufacturer’s instruction, and genotypically by random amplification of polymorphic DNA (RAPD) typing, using short primer sequence (Table 1; [16]).
Testing the phenotypic and genotypic changes in the generated mutants
Cross-resistance to different antibiotics
For the parent strain as well as the resultant mutants, the MIC values were determined against selected group of antimicrobial agents (cefepime, ceftazidime, meropenem, ciprofloxacin, amikacin, and colistin), by broth microdilution method, as described previously in determination of CHX MIC. Each antimicrobial agent was tested in a concentration range from 0.0625 to 128 µg/mL. The results of MIC were interpreted as sensitive, intermediate, and resistant following clinical and laboratory standards institute guidelines [19].
Change in membrane permeability
The possible changes in the membrane permeability of the mutant strains, relative to that of the parent strain, was assessed using the fluorescent probe, 8-anilino-1-naphthylenesulfonic acid (ANS, Sigma-Aldrich, Germany); ANS is a neutrally charged hydrophobic probe that fluoresces weakly in aqueous environments and exhibits enhanced fluorescence in hydrophobic environments [20]. Relative fluorescence was calculated as the ratio of ANS fluorescence in mutant cells to that in the parent P. aeruginosa PAO1 cells. The assay was performed in independent triplicates with each biological replicate consisting of three technical replicates.
Changes in outer membrane proteins
Outer membrane vesicles (OMV) were obtained from the culture of parent P. aeruginosa PAO1 strain as well as the tested mutants using the ethylenediaminetetraacetic acid (EDTA)–based method described by Murphy and Loeb [21] with slight modifications [22]. Briefly, overnight bacterial cells were spun down by centrifugation at 6000 × g for 30 min and re-suspended in 15 mL of EDTA buffer. The collected bacterial cells in EDTA were transferred to a 100-mL flask whose bottom was covered with 3-mm glass beads and shaken vigorously at 250 rpm for 45 min at 55 °C. Cell debris was removed by centrifugation at 10,000 × g for 15 min at 4 °C and the OMV were obtained by centrifugation of the supernatant at 22,000 × g for 90 min at 4 °C. Membrane vesicles in the pellet were re-suspended in 150 µL cold phosphate buffered saline.
Protein content of OMV was determined using Bradford assay [23] and sample volumes equivalent to 10 µg of outer membrane proteins were further analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The SDS-PAGE gel was prepared using separating gel concentration of 17.5%.
Detection of efflux-mediated resistance
Chlorpromazine (CPZ; Sigma-Aldrich, Germany) was used, as an efflux pump inhibitor, for detection of presence of efflux-mediated resistance [24]. To confirm the absence of any antimicrobial activity of CPZ, its MIC was determined by broth microdilution method, as described previously. CPZ was tested in a concentration range from 12.5 to 400 µg/mL, against the parent P. aeruginosa PAO1 strain and the generated mutants. The MIC values of CHX were determined in absence and presence of 100 µg/mL of CPZ, against the parent P. aeruginosa PAO1 strain and the generated mutants, using broth microdilution method.
Effect of exposure to sub-inhibitory concentration of CHX on expression of P. aeruginosa multidrug efflux pumps
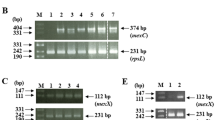
The effect of exposure to sub-inhibitory concentrations of CHX on the level of expression of P. aeruginosa PAO1 multidrug efflux pumps MexAB-oprM, MexEF-oprN, and MexXY was tested by real-time reverse transcription-polymerase chain reaction (RT-PCR). An overnight culture, of P. aeruginosa PAO1 strain, was diluted 1:100 in 50 mL MH broth, with and without 0.5 MIC of CHX (0.781 µg/mL), representing test and standard conditions, respectively. The broth tubes were incubated at 37 °C with shaking at 180 rpm. RNA was extracted, at logarithmic phase (OD600 = 0.45 ± 0.05), using the RNeasy Mini Kit (Qiagen, Germany), according to the manufacturer’s instructions.
Complementary DNA (cDNA) was prepared using Quantitect reverse transcription kit (Qiagen, Germany), according to the manufacturer’s instructions. Two negative controls were included; a no template control (NTC), without RNA template to serve as a general control for extraneous nucleic acid contamination, and a no reverse transcriptase control (NRT) to assess the presence of any DNA contamination in the RNA preparation.
The concentration and purity, of the resultant cDNA, were evaluated using nanophotometer (IMPLEN GmbH, Germany). The expression of the genes for the membrane fusion proteins mexA, mexE, and mexX was used to represent the expression of the efflux pumps MexAB-oprM, MexEF-oprN, and MexXY, respectively. rpsL gene was used as a house keeping gene to normalize the results. The reaction primers were designed using integrated DNA technologies primer quest tool (https://eu.idtdna.com/primerquest/home/index). All primers (Macrogen Inc., South Korea) were designed to have an annealing temperature of 62 °C and expected amplicon sizes of nearly 100 bp (Table 1).
The real-time RT-PCR reaction was carried out in a Rotor-Gene Q real-time PCR cycler (Qiagen, Germany) using GoTaq® qPCR Master Mix (Promega, USA), following the reaction conditions and instructions recommended by the manufacturer. Each sample was tested in duplicates. NRT control was used to guard against any possible DNA contamination.
Determination of MexXY efflux pump expression level in CHX-resistant mutants
The level of mexX gene expression in the generated CHX-resistant mutants was determined and compared to that in the parent P. aeruginosa PAO1 strain, using real-time RT-PCR as described above.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.01 (GraphPad software Inc., CA, USA). A p-value ≤ 0.05 was considered significant. The results of mutation frequency, mutation rate, and growth rate experiments were compared using the Kruskal–Wallis test. Results of RT-PCR were compared using the paired student t-test, while one-sided unpaired t test was used to compare the relative fluorescence (mutant/parent) of the mutants to that of the parent strain, to test presence of significant reduction in permeability that might be responsible for CHX resistance. The pairwise correlation between fold increase in mutants' MICs of CHX and of tested antibiotics was explored using Spearman’s correlation coefficient.
Results
CHX minimum inhibitory concentration
The MIC of CHX against the standard P. aeruginosa PAO1 strain was 1.56 µg/mL as determined by broth microdilution method.
Mutation frequency and mutation rate after spontaneous and short-term exposure to CHX
The spontaneous mutation frequency after exposure to lethal concentration of CHX (32 times the MIC; 50 µg/mL) and the mutation frequencies after short-term exposure to sub-inhibitory concentrations of CHX (0.125, 0.25, and 0.5 of the MIC) were evaluated through determination of the resistance index in each case. The resistance index (the mean of the number of resistant cells divided by the total number of viable cells per culture) was calculated in three independent biological replicates. Using the Kruskal–Wallis test, no significant difference (p > 0.99) was detected between the resistance index determined after spontaneous exposure to CHX (mean of resistance index ± standard deviation was 4.7 × 10−7 ± 5.2 × 10−7) and after short-term exposure to sub-inhibitory concentrations of CHX ( mean of resistance index ± standard deviation were 8.8 × 10−7 ± 6.8 × 10−7, 1.1 × 10−6 ± 1.2 × 10−6, and 4.99 × 10−7 ± 4.5 × 10−7 after exposure to 0.125, 0.25, and 0.5 of the CHX MIC, respectively). Similarly, no significant difference (p > 0.05) was recorded between the mutation rates calculated after spontaneous exposure to CHX (mean of mutation rate ± standard deviation was 1.55 × 10−7 ± 1.2 × 10−6) and after short-term exposure to sub-inhibitory concentrations of CHX (mean of mutation rates ± standard deviation were 7.9 × 10−7 ± 6.2 × 10−7, 1.4 × 10−6 ± 1.4 × 10−6, and 5.5 × 10−7 ± 4.9 × 10−7 after exposure to 0.125, 0.25, and 0.5 of the CHX MIC, respectively).
Growth of P. aeruginosa PAO1 strain in presence of sub-inhibitory concentrations of CHX
The effect of presence of sub-inhibitory concentrations of CHX on the growth pattern of P. aeruginosa PAO1 strain was monitored by measuring the optical density, of the growth culture, at 600 nm for a period of 24 h (until reaching the stationary phase). Similar growth patterns were observed in the presence of 0.25 and 0.5 CHX MIC and in absence of CHX, regardless of its addition from the start of the incubation period (at time = 0 h) or after 5 h of incubation (Fig. 1). In addition, calculation of growth rate confirmed the lack of any effect of CHX addition on the growth rate of P. aeruginosa (p > 0.05 using Kruskal–Wallis test). A mean growth rate ± standard deviation was determined to be 0.42 ± 0.11 h−1, 0.4567 ± 0.12 h−1, and 0.431 ± 0.04 h−1 for culture without CHX, with 0.25 of CHX MIC and for culture with 0.5 of CHX MIC, respectively. It was not practical to calculate growth rates for cultures with CHX added at t = 5 h; however, these cultures had superimposed growth pattern to that of other cultures (Fig. 1).
Generation of CHX-resistant mutants
Serial passage of P. aeruginosa PAO1 strain in increasing concentrations of CHX generated 18 different resistant phenotypes that were able to grow at the higher CHX concentrations than the MIC of the parent strain, for three successive passages. In addition, ten resistant colonies were picked randomly from those exposed to CHX (32 times the MIC), either spontaneously or after short-term exposure to sub-inhibitory concentrations of CHX (mutation frequency plates).
Only twelve out of the 28 resistant phenotypes were stable after serial cultivation overnight in MH broth without CHX; these mutants had CHX MIC more than or equal to eight-fold (≥ 12.5 µg/mL) the MIC of the parent P. aeruginosa PAO1 (1.56 µg/mL; Table 2). The MIC of each resistant phenotype after passage in increasing CHX concentrations and the MIC of the corresponding mutants after passage in MH broth without CHX are indicated in Fig. 2.
Minimum inhibitory concentration (MIC) of chlorhexidine (CHX)–resistant phenotypes and CHX-resistant mutants generated by serial passage in increasing CHX concentrations. The circles on the black line represent the resistant phenotypes isolated after each passage sequentially (P1 to P18 from passages 1–18). The triangles on the red line represent the corresponding mutants after passage in CHX-free Muller Hinton broth sequentially (M1 to M9 for mutants with MIC ≥ 12.5 µg/mL). Dotted horizontal line represents the cutoff value for selection of resistant mutants that had maintained MIC ≥ eight-fold (12.5 µg/mL) that of parent P. aeruginosa PAO1 strain after passage in CHX-free broth
Confirmation of the purity of the mutant strains
The purity of the mutant strains was confirmed phenotypically using API 20 NE identification system. All the mutants produced an API 20 NE profile identical (99.9%) to that of parent strain (P. aeruginosa PAO1). Their purity was also confirmed genotypically using RAPD Typing; the band patterns of the stable resistant mutants were similar to that of the parent strain.
Cross-resistance of the generated mutants to different antibiotics
The MICs of different antibiotics (amikacin, ciprofloxacin, meropenem, ceftazidime, colistin, and cefepime) against the parent strain and the generated mutants were determined by broth microdilution method. All mutants had a two to fourfold increase in MIC values against amikacin while only some of them had an increase in MIC against ciprofloxacin, meropenem, ceftazidime, colistin, and cefepime (Table 3). The fold change in MIC against cefepime and meropenem was partially and significantly correlated with the fold change in MIC of CHX (0.58 and 0.83, respectively; p < 0.05).
Several mutants shifted to a higher resistance level against some of the tested antibiotics. The mutants M3, M7, and M8 changed their pattern from sensitive to intermediate resistance against cefepime while the mutants M3, M4, M6, M7, M8, and M9 changed their pattern from sensitive to intermediate in case of ciprofloxacin. With meropenem, several mutants became resistant: M2, M3, M4, M5, M7, M8, and M12. For amikacin, ceftazidime, and colistin antibiotics, all mutants remained sensitive although there was a reported increase in their MIC values.
Change in membrane permeability
The changes in membrane permeability of the mutant strains, relative to that of the parent strain, were evaluated using the fluorescent probe ANS. The relative fluorescence was calculated as the ratio of ANS fluorescence in mutant cells to that in the parent P. aeruginosa PAO1 cells. Several mutants showed a decrease in relative fluorescence and hence the membrane permeability, while others showed increase in the relative fluorescence. Applying one-sided unpaired t test, in comparing the relative fluorescence of the mutants (mutant/parent), only M8 and M12 showed a significant decrease in relative florescence (p < 0.05; Fig. 3).
Changes in the outer membrane protein
The OMP were separated from the parent strain (P. aeruginosa PAO1) and all mutants. They were compared by visualization using SDS-PAGE. All mutants, except M12, showed a protein band at 20 KDa that was not detectable in the parent strain. In addition, two proteins of about 35 KDa disappeared in M4 and M7 mutants compared to the parent strain, while showing reduced detectability in the remaining strains (Fig. 4).
Detectability of efflux-mediated resistance
CPZ was used as efflux pump inhibitor. Its antimicrobial activity against the parent strain (P. aeruginosa PAO1) and the tested mutants was assessed by determination of its MIC; CPZ had no antimicrobial activity below 250 µg/mL (MIC = 400 µg/mL). The presence of efflux-mediated resistance to CHX was tested by determination of CHX MIC, for the parent strain and the tested mutants, in the presence and absence of CPZ. In the presence of 100 µg/mL CPZ, all mutants showed ≥ four-fold decrease in their CHX MIC. CPZ was also able to reverse CHX resistance phenotype for all tested mutants except M8 and M9 (Table 4). In M6 and M7, partial reversion in resistance toward CHX was recorded where the MIC of CHX in the presence of CPZ was equal to that of the CHX MIC against the parent P. aeruginosa PAO1 strain in absence of CPZ.
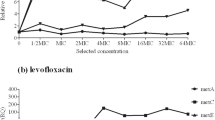
Effect of short-term exposure to sub-inhibitory concentration of CHX on the expression of selected efflux pumps
The expression of the selected efflux pump genes (mexA, mexE, and mexX) in presence of 0.5 value of CHX MIC (0.781 µg/mL) was determined in the parent strain P. aeruginosa PAO1 as fold change relative to the expression in absence of CHX, by real-time RT-PCR. Presence of CHX at concentration equal to 0.5 of MIC significantly upregulated the expression of mexX gene. However, the expression of mexA and mexE genes did not change significantly (Fig. 5).
Determination of the level of expression of MexXY efflux pump in the generated mutants
The level of expression of mexX gene was determined in the generated mutants and was compared to its level of expression in the parent strain, as indicative of MexXY efflux pump expression. All of the mutants except M6 and M9 recorded a higher level of expression of mexX gene compared to that in parent P. aeruginosa PAO1 with seven out of 12 mutants had a significant level of overexpression that reached about 43-fold (M1, M3, M4, M5, M7, M11, and M12) compared to the parent strain (p < 0.0001; Fig. 6).
Discussion
In this study, the possible effect of CHX exposure, on the generation of resistant mutants in P. aeruginosa PAO1 strain, was tested. No significant difference was detected, in the mutation frequency or the mutation rate, after short-term exposure to different sub-MICs of CHX. However, some stable resistant mutants were generated and used for further testing. In a similar observation, the presence of triclosan did not increase the mutation frequency of Salmonella enterica serovar Typhimurium but was able to select bacterial strains with increased antibiotic resistance [16].
Different methods can be used to generate the adapted resistant strains, depending on the concentration of the agent exerting the selective pressure, whether it is applied in lethal or sub-lethal concentration, and the repetition of exposure whether single or repeated [25]. In this study, resistant strains were generated through two different models; continuous exposure by serial passage in increasing concentrations of CHX and single exposure to lethal CHX concentration, by randomly selecting the resistant colonies that grew in 32 times the value of CHX MIC (50 mg/L).
Growth of P. aeruginosa in gradually increasing CHX concentration resulted in generation of strains with increased CHX MIC. Similar to our results, the increase in chlorhexidine MIC, by serial passage in gradually increasing CHX concentrations, was reported previously in about 45 bacterial species including P. aeruginosa. Also, some of these studies reported an increase in CHX MIC in P. aeruginosa after passage in fixed sub-inhibitory CHX concentration [25, 26].
To rule out the unstable resistance phenotypes, the generated resistant phenotypes were subjected to serial passage in plain MH broth. Finally, 12 stable mutants were generated with eight to 16-fold increase in CHX MIC. The generation of stable CHX resistance, in P. aeruginosa, following exposure to gradually increasing concentrations of CHX was reported previously [25, 27]. However, the resistant P. aeruginosa mutants generated by Forbes et al. and Tattawasart et al. rapidly lost their CHX resistance [28, 29]. The development of stable CHX resistance in Klebsiella pneumoniae and Enterococcus faecium has been recorded [30, 31].
The generated stable mutants have shifted to a higher MIC level (two to four-fold) against some of the tested antibiotics (cefepime, ceftazidime, meropenem, amikacin, colistin, and ciprofloxacin). The increase in MIC of meropenem and cefepime was significant and partially correlated with CHX MIC increase in the generated mutants, indicating the possible involvement of common resistance mechanism. It is highly worrisome that seven out of the 12 generated mutants gained meropenem resistance by chlorhexidine exposure; the World Health Organization considered carbapenem-resistant P. aeruginosa among the critical list of pathogens for development of new antimicrobials [12]. Carbapenems are among the most active and potent agents against MDR Gram-negative bacteria and development of resistance to carbapenems leaves few therapeutic options [32]. This cross-resistance, between the antibiotics and CHX, is consistent with the reported decrease in antibiotic susceptibility of chlorhexidine-resistant Gram-negative bacteria [33]. However, Thomas and colleagues reported preservation of the susceptibility, to different antibiotics, in the generated CHX-resistant P. aeruginosa strains [25].
No cross-resistance to colistin antibiotic was detectable; conversely, other studies have reported an association between CHX adaptation and development of colistin resistance in P. aeruginosa and K. pneumoniae [27]. This variation in detectability of colistin resistance among CHX-resistant strains may have resulted from different non-synonymous mutations and consequently different proteomic changes. Further studies are therefore required to elucidate the possible mutations responsible for the development of colistin resistance in CHX-resistant mutants.
Although the resistant mutants were generated by exposure to CHX concentrations ranging between 0.78 and 50 µg/mL which are lower concentrations than those used in pharmaceutical CHX products, however, low concentrations of CHX may remain on the skin after use and select for CHX resistance. In addition, some eye preparations contain 50 mg/L of CHX as a preservative [34] which may select CHX-resistant mutants that are able to contaminate such preparations. Contamination of CHX-containing preparations by P. aeruginosa was reported previously [33]. This generation of CHX-resistant mutants with antibiotics cross-resistance is highly worrying, especially in hospital environment, where it was reported recently that CHX-resistant P. aeruginosa cells have proteomic changes promoting their tissue colonization and virulence [27].
To evaluate the impact of CHX exposure on the emergence of antibiotic resistance, it was important to understand the possible mechanisms that contribute to such phenotype. The detected cross-resistance to antimicrobial agents from different classes suggested the possibility of non-specific mechanisms of resistance. Decreased accumulation of the drug is an important mechanism of resistance in many clinical isolates of P. aeruginosa. This type of resistance arises from either restricted outer membrane permeability, and/or drug efflux [11, 35].
Two of the generated mutants (M8 and M12) showed a statistically significant decrease in membrane permeability compared to the parent strain. This could account for their CHX resistance, where CHX enters the cell through simple diffusion that is self-promoted by the compound itself [36]. This decrease in permeability could have arisen from the upregulation of several membrane proteins, such as LptD, MurD, and PagL, following CHX exposure [27]. The permeability decrease, in the generated P. aeruginosa mutants, could not explain their increased resistance to some antibiotics as cefepime, meropenem, and ciprofloxacin; these antibiotics use porin for their penetration [35]. In this case, other mechanisms of resistance, such as efflux pumps or porin modification, may account for antibiotic resistance. The absence of decreased membrane permeability in mutants other than M8 and M12 suggested the possibility of other CHX resistance mechanisms than reduced membrane permeability.
Analyzing the OMP profile, of the generated resistant mutants, revealed loss of two OMP of about 35 KDa in M4 and M7 mutants, compared to the parent strain. Furthermore, the detectability of these two proteins was reduced in the rest of mutants where OMP may function as a porin, facilitating the uptake of antibiotics such as meropenem, ceftazidime, and cefepime. The absence of the outer membrane protein T-OMP of similar size (35 KDa) was recorded to affect the onset of antimicrobial susceptibility in P. aeruginosa [37]. In addition, the effect of porin loss, on the resistance to several antimicrobial compounds in P. aeruginosa strains, was reported previously [35, 38]; however, no studies are available on the role of porins in CHX resistance.
All mutants, except M12, showed a protein band at nearly 20 KDa that was not detectable in the parent strain. Presence of a new OMP in resistant mutants is diagnostic for MDR systems [39]; these proteins can represent a component of an overexpressed efflux pump. Tabata and colleagues reported a significant overexpression of OprR protein (approximately 26 kDa) in mutants adapted to quaternary ammonium compounds [40]. CHX also induces the expression of OprH, an outer membrane protein of approximately 20 kDa, in P. aeruginosa [41]. The overexpression of the OMP H1 precursor (OprH) limits the transportation of cationic antimicrobial agents as aminoglycosides and polymyxins [42]. The self-promoted uptake of CHX, being a cationic agent, can also be reduced by the overexpression of this OMP which needs further investigation.
Although several researchers have denied the role of efflux in chlorhexidine resistance [43], however, there is several evidences that efflux pumps could explain decreased susceptibility to CHX as well as to other antibiotics, in P. aeruginosa strains [44]. In this study, CPZ was able to reverse or reduce CHX resistance phenotype for all tested mutants. This confirmed the role of efflux in CHX resistance. Reduction in CHX MIC in P. aeruginosa and K. pneumoniae, by the presence of efflux pump inhibitors, was reported previously [10, 45]. This efflux-mediated resistance may be responsible for cross-resistance to different antimicrobial agents [11].
Exposure of P. aeruginosa to sub-inhibitory concentrations of some antimicrobial agents upregulates the expression of the MDR efflux systems, Mex [41]. In this study, exposure to 0.5 the value of CHX MIC upregulated the expression of the MexXY multidrug efflux pump. In addition, nearly half of CHX-resistant mutants recorded a significantly increased level of MexXY efflux pump expression (seven out of 12) indicating the role of CHX in selection of P. aeruginosa–resistant strains overexpressing MexXY efflux system. To the best of our knowledge, this is the first report on the role of CHX in induction or selection of MexXY efflux pump overexpression. Previous proteomics analysis on P. aeruginosa, exposed to serial passage of CHX, reported upregulation of multidrug-resistance protein MexA, the periplasmic linker component of the MexAB-OprM efflux system that confers multidrug resistance [27]. Also, CHX-resistant environmental isolates with MexAB overexpression and cross-resistance to different antibiotics were recently reported [46]. Fluoroquinolones, β-lactams, tetracyclines, aminoglycosides, macrolides, and chloramphenicol are substrates for MexXY efflux pump [47]. Overexpression of MexXY efflux pump was reported to be responsible for resistance to different antimicrobial agents as aminoglycosides, fluoroquinolone, and cefepime [47, 48]. Hence, CHX may have contributed to the reduced antibiotic susceptibility.
This is a preliminary study to detect the possible phenotypic changes associated with CHX resistance and their role in antibiotic resistance. However, these detected changes need to be fully and specifically elucidated due to the complex nature of the detected changes and the complex response of P. aeruginosa to various antimicrobial agents. Further studies are still required to determine the specific type of the protein that was induced or disappeared from the OMP of CHX-resistant mutants and their possible role in CHX and antimicrobial resistance. The level of OprM protein needs to be determined in MexXY overexpressing mutants, where OprM protein is necessary for MexXY functioning. Similarly, the effect of CPZ on efflux pump expression and the specific type of efflux pump affected by CPZ needs further studies, where about 12 resistance-nodulation-division efflux pumps were reported to be implicated in P. aeruginosa PAO1 antimicrobial resistance. Fewer number of efflux pumps from other families were also found to play a role in antimicrobial resistance [49]. The possible overexpression of efflux systems other than MexAB-OprM or MexXY in CHX-resistant mutants and their possible role in cross-resistance to different antibiotics need further investigation.
Conclusion
Exposure to CHX, either single or repeated, has a selective pressure for resistant P. aeruginosa strains with cross-resistance to antibiotics from different classes. This can be mediated by different phenotypic and genotypic mechanisms. Therefore, special protocols are required to be implemented during the use of CHX containing preparations; the benefit versus risk for personal-care products containing CHX needs further assessment.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Centers for Disease Control and Prevention. 2019 AR threats report [Internet]. 2019; https://www.cdc.gov/drugresistance/biggest-threats.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fdrugresistance%2Fbiggest_threats.html
Willyard C (2017) The drug-resistant bacteria that pose the greatest health threats. Nat News 543:15
Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP (2001) Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother. 45:428–432. https://www.ncbi.nlm.nih.gov/pubmed/11158736: https://doi.org/10.1128/AAC.45.2.428-432.2001
Morita Y, Murata T, Mima T, Shiota S, Kuroda T, Mizushima T et al (2003) Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother 51:991–994. https://doi.org/10.1093/jac/dkg173
Slipski CJ, Zhanel GG, Bay DC (2018) Biocide selective TolC-independent efflux pumps in Enterobacteriaceae. J Membr Biol 251:15–33. https://doi.org/10.1007/s00232-017-9992-8
FDA issues final rule on safety and effectiveness of antibacterial soaps | FDA. FDA (2016) [cited 2019 24]; https://www.fda.gov/news-events/press-announcements/fda-issues-final-rule-safety-and-effectiveness-antibacterial-soaps. Accessed 29 May 2017
Horner C, Mawer D, Wilcox M (2012) Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 67:2547–2559. https://doi.org/10.1093/jac/dks284
Stickler DJ, Thomas B (1980) Antiseptic and antibiotic resistance in Gram-negative bacteria causing urinary tract infection. J. Clin Pathol. 33:288–96. https://www.ncbi.nlm.nih.gov/pubmed/6769972https://doi.org/10.1136/jcp.33.3.288
Stein C, Vincze S, Kipp F, Makarewicz O, Al Dahouk S, Pletz MW (2019) Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect Dis 19:932–933. https://doi.org/10.1016/S1473-3099(19)30427-X
Wand ME, Bock LJ, Bonney LC, Sutton JM (2017) Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother 61:1–31. https://doi.org/10.1128/AAC.01162-16
Lister PD, Wolter DJ, Hanson ND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. https://doi.org/10.1128/CMR.00040-09
World Health Organization (2017) WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. Accessed March 2018
Ostroff RM, Wretlind B, Vasil ML (1989) Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect Immun 57:1369–1373
Wayne, PA (2015) Clinical and Laboratory Standards Institute. M07-A10: methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved Standard—Tenth Edition
Tsuchiya K, Cao Y-Y, Kurokawa M, Ashino K, Yomo T, Ying B-W (2018) A decay effect of the growth rate associated with genome reduction in Escherichia coli. BMC Microbiol 18:101. https://doi.org/10.1186/s12866-018-1242-410.1186/s12866-018-1242-4
Birosova L, Mikulasova M (2009) Development of triclosan and antibiotic resistance in Salmonella enterica serovar Typhimurium. J Med Microbiol. 58:436–41. http://jmm.microbiologyresearch.org/content/journal/jmm/https://doi.org/10.1099/jmm.0.003657-0
Rosche WA, Foster PL (2000) Determining mutation rates in bacterial populations. Methods 20:4–17. https://doi.org/10.1006/meth.1999.0901
Lanini S, D’Arezzo S, Puro V, Martini L, Imperi F, Piselli P, et al (2011) Molecular epidemiology of a Pseudomonas aeruginosa hospital outbreak driven by a contaminated disinfectant-soap dispenser. PLoS One. 6. https://doi.org/10.1371/journal.pone.0017064
Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing; Twenty-Seventh Informational Supplement. CLSI Document M100-S27. www.clsi.org
Lamers RP, Cavallari JF, Burrows LL (2013) The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 8:1–7. https://doi.org/10.1371/journal.pone.0060666
Murphy TF, Loeb MR (1989) Isolation of the outer membrane of Branhamella catarrhalis. Microb Pathog 6:159–174. https://doi.org/10.1016/0882-4010(89)90066-1
Mohamed SA (2015) A study of the role of interaction between Moraxella catarrhalis surface antigen and specific host antibody response on the occurance of otitis media. Master thesis, Cairo University
Bradford MM (1976) A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Kristiansen JE, Thomsen VF, Martins A, Viveiros M, Amaral L (2010) Non-antibiotics reverse resistance of bacteria to antibiotics. In Vivo (Brooklyn) 24:751–754
Thomas L, Maillard J, Lambert RJW, Russell AD (2000) Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a ‘ residual ’ concentration. 297–303. https://doi.org/10.1053/jhin.2000.0851
Kampf G (2019) Adaptive bacterial response to low level chlorhexidine exposure and its implications for hand hygiene. Microb. cell (Graz, Austria) 6:307–320. https://doi.org/10.15698/mic2019.07.683
Hashemi MM, Holden BS, Coburn J, Taylor MF, Weber S, Hilton B, et al (2019) Proteomic analysis of resistance of Gram-negative bacteria to chlorhexidine and impacts on susceptibility to colistin , antimicrobial peptides, and ceragenins. 10:1–13. https://doi.org/10.3389/fmicb.2019.00210
Forbes S, Dobson CB, Humphreys GJ, McBain AJ (2014) Transient and sustained bacterial adaptation following repeated sublethal exposure to microbicides and a novel human antimicrobial peptide. Antimicrob Agents Chemother. 58:5809–17. https://www.ncbi.nlm.nih.gov/pubmed/25049246https://doi.org/10.1128/AAC.03364-14
Tattawasart U, Maillard JY, Furr JR, Russell AD (1999) Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J Hosp Infect 42:219–229. https://doi.org/10.1053/jhin.1999.0591
Bock LJ, Wand ME, Sutton JM (2016) Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect 93:42–48. https://doi.org/10.1016/j.jhin.2015.12.019
Bhardwaj P, Hans A, Ruikar K, Guan Z, Palmer KL (2017) Reduced chlorhexidine and daptomycin susceptibility in vancomycin-resistant Enterococcus faecium after serial chlorhexidine exposure. Antimicrob. Agents Chemother 62: e01235–17. https://doi.org/10.1128/AAC.01235-17
Doi Y (2019) Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin Infect Dis 69:S565-S575. https://doi.org/10.1093/cid/ciz830
Kampf G (2018) Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 7:110. https://doi.org/10.3390/antibiotics7040110
Kramer A, Behrens-Baumann W (2002) Antiseptic prophylaxis and therapy in ocular infections: principles, clinical practice and infection control. Karger Medical and Scientific Publishers
Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. https://doi.org/10.1016/j.bbapap.2008.11.005
Denyer SP, Maillard J-Y (2002) Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. J Appl Microbiol 92(Suppl):35S-45S
Brozel VS, Cloete TE (1994) Resistance of Pseudomonas aeruginosa to isothiazolone. J Appl Bacteriol 76:576–582
Yamano Y, Nishikawa T, Komatsu Y (1990) Outer membrane proteins responsible for the penetration of beta-lactams and quinolones in Pseudomonas aeruginosa. J Antimicrob Chemother 26:175–184. https://doi.org/10.1093/jac/26.2.175
Alonso A, Campanario E, Martínez JL (1999) Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145:2857–2862. https://doi.org/10.1099/00221287-145-10-2857
Tabata A, Nagamune H, Maeda T, Murakami K, Miyake Y, Kourai H (2003) Correlation between resistance of Pseudomonas aeruginosa to quaternary ammonium compounds and expression of outer membrane protein OprR 1. Antimicrob. Agents Chemother. 47:2093–9. isi:000183786700005 https://doi.org/10.1128/AAC.47.7.2093
Morita Y, Tomida J, Kawamura Y (2014) Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 4:422. https://doi.org/10.3389/fmicb.2013.00422
Young ML, Bains M, Bell A, Hancock RE (1992) Role of Pseudomonas aeruginosa outer membrane protein OprH in polymyxin and gentamicin resistance: isolation of an OprH-deficient mutant by gene replacement techniques. Antimicrob Agents Chemother 36:2566–2568. https://doi.org/10.1128/aac.36.11.2566
Gilbert P, Moore LE (2005) Cationic antiseptics: diversity of action under a common epithet. J Appl Microbiol 99:703–715. https://doi.org/10.1111/j.1365-2672.2005.02664.x
Poole K (2005) Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51. https://doi.org/10.1093/jac/dki171
Mombeshora M, Mukanganyama S (2017) Development of an accumulation assay and evaluation of the effects of efflux pump inhibitors on the retention of chlorhexidine digluconate in Pseudomonas aeruginosa and Staphylococcus aureus. BMC Res Notes 10:1–9. https://doi.org/10.1186/s13104-017-2637-2
Amsalu A, Sapula SA, De Barros Lopes M, Hart BJ, Nguyen AH, Drigo B, et al (2020) Efflux pump-driven antibiotic and biocide cross-resistance in Pseudomonas aeruginosa isolated from different ecological niches: a case study in the development of multidrug resistance in environmental hotspots. Microorganisms. 24;8:1647. https://pubmed.ncbi.nlm.nih.gov/33114277https://doi.org/10.3390/microorganisms8111647
Aeschlimann JR (2003) The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 23:916–24
Hocquet D, Muller A, Blanc K, Plésiat P, Talon D, Monnet DL et al (2008) Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1173–1175. https://doi.org/10.1128/AAC.01212-07
Li X-Z, Plésiat P (2016) Antimicrobial drug efflux pumps in Pseudomonas aeruginosa. In: Li X-Z, Elkins CA, Zgurskaya HI, editors. Efflux-mediated antimicrobial resistance in bacteria. 1st ed. Cham: Springer International Publishing. pp. 359–400. https://doi.org/10.1007/978-3-319-39658-3_14
Acknowledgements
The authors would like to thank Dr. Ahmed Sherif Attia (Cairo University) for providing P. aeruginosa PAO1 strain, Dr. Mohamed Abdel Halim Ramadan (Cairo University) for providing chlorhexidine hydrochloride, and Dr. Shahira Abdel Salam (Cairo University) for her technical assistance.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.S.Y., O.E., and M.T.K.; methodology, M.A.T; formal analysis, M.A.T., A.S.Y., and M.T.K.; writing—original draft preparation, M.A.T.; writing—review and editing, A.S.Y. and M.T.K.; supervision, A.S.Y. and M.T.K. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the ethics committee of Faculty of Pharmacy, Cairo University (approval number: MI 1617).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tag ElDein, M.A., Yassin, A.S., El-Tayeb, O. et al. Chlorhexidine leads to the evolution of antibiotic-resistant Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 40, 2349–2361 (2021). https://doi.org/10.1007/s10096-021-04292-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04292-5