Abstract
The use of molecular assays to rapidly identify pathogens and resistance genes directly from positive blood cultures (BCs) contribute to shortening the time required for the diagnosis of bloodstream infections. In this work, loop-mediated isothermal amplification (LAMP) assays have been examined for their potential use in BC diagnosis. Three different assays were applied. The commercially available eazyplex® MRSA test detects Staphylococcus aureus, S. epidermidis, mecA, and mecC. Two in-house assays [Gram-positive (GP) and Gram-negative (GN)] have been developed for the detection of streptococci, enterococci, vanA, vanB, Pseudomonas spp., Enterobacteriaceae, and the bla CTX-M family. A total of 370 positive BCs were analyzed. LAMP test results were obtained within 30 min, including sample preparation. Amplification was measured by real-time fluorescence detection. The threshold time for fluorescence intensity values ranged from 6.25 to 13.75 min. The specificity and sensitivity of the assays varied depending on the target. Overall, from 87.7% of BCs, true-positive results were obtained, compared to routine standard diagnosis. Twenty-one tests were true-negative because of the lack of an appropriate target (5.7%). The concordance of positive test results for resistance genes with subsequent antibiotic susceptibility testing was 100%. From 15 BC bottles with mixed cultures, eazyplex® assays produced correct results in 73% of the cases. This study shows that LAMP assays are fast and cost-saving tools for rapid BC testing in order to expedite the diagnostic report and improve the antibiotic stewardship for sepsis patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Time-saving microbiological diagnosis of sepsis remains a key challenge for the clinical laboratory. The initial use of inappropriate antibiotics increases the mortality of patients with sepsis, underlining that an early targeting of the antibiotic therapy is highly important for an improved patient care [1]. Bloodstream infections can be diagnosed by classical blood culture (BC) and culture-independent polymerase chain reaction (PCR)-based approaches. PCR conducted directly on patient blood samples can basically produce faster results and avoids the problem of high contamination rates of BCs with coagulase-negative staphylococci (CoNS) [2]. However, labor-intensive PCR assays are difficult to integrate into a continuous routine lab flow. A recently introduced commercial assay based on PCR/electrospray ionization-mass spectrometry (PCR/ESI-MS) offers reduced hands-on time but requires high investment costs and has only a limited capacity of samples that can be analyzed during a normal working day [3]. One major problem of BC diagnosis, the delay in reporting results due to conventional subculture for identification and antimicrobial susceptibility testing (AST), can be avoided by the application of rapid molecular tests or matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) to identify pathogens and their key resistance markers directly from positive BC bottles [4–6]. Automated random access tests have been shown to be easily applicable as reliable tools for BC diagnosis, providing results within one or two hours [7, 8]. Such assays with a minimum of hands-on time can contribute to implement a targeted antibiotic therapy as soon as possible [9]. The choice of an appropriate test for the routine lab depends on costs for machines and consumables, test performance, and an easy handling, allowing integration into the daily routine workflow. An alternative technology for rapid molecular assays is provided by loop-mediated isothermal amplification (LAMP), an amplification technique that uses Bst DNA polymerase with strand-displacing activity [10]. LAMP offers high-speed amplification within several minutes under isothermal conditions and does not require DNA purification from most clinical sample types [11, 12]. Its reliability to detect carbapenemases and extended-spectrum beta-lactamase (ESBL) genes in cultured bacteria and directly from urine samples of patients with urinary tract infection has been recently demonstrated [13–15].
This study was designed to evaluate the potential of LAMP to rapidly identify the most common pathogens and their major resistance genes from positive BC bottles within a time frame of 30 min. Three different eazyplex® LAMP assays with real-time fluorescence detection of amplificates were applied. The eazyplex® MRSA (Amplex BioSystems, Giessen, Germany) is a CE-labeled commercial test for the identification of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) from nasal and pharyngeal swabs and was used with a preliminary BC protocol. The eazyplex® Gram-positive (GP) and Gram-negative (GN) assays were developed for study purposes to detect streptococci, enterococci, and Enterobacteriaceae and Pseudomonas, respectively. All LAMP assays were evaluated using routine BCs as clinical samples.
Materials and methods
Bacterial strains and in-house LAMP assays
Specific primer sets were designed using LAMP Designer software v1.10 (PREMIER Biosoft International, Palo Alto, CA, USA). For the different species or genera, the following target genes were chosen for primer design: GP assay: Enterococcus spp. (tufA), E. faecalis (EF0027, coding for a phosphosugar-binding transcriptional regulator protein), Streptococcus pneumoniae (lytA), Streptococcus spp. (tufA); GN assay: Escherichia coli (yfiL), Klebsiella pneumoniae (ydhS), Pseudomonas aeruginosa (oprL) and 16s rDNA for Enterobacteriaceae (Table 1). A total of 19 Gram-positive and 64 Gram-negative isolates and reference strains purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) or obtained from the National Reference Laboratory for Multidrug Resistant Gram-negative Bacteria (Bochum, Germany) were used to evaluate the specificity of the primer sets of the in-house GP and GN LAMP assays (Table 1). A small single colony was suspended in 500 μL of resuspension and lysis fluid (RALF, Amplex BioSystems) and boiled for 2 min. 25 μL of the suspension were added to each of the tubes in the eazyplex® test strip. The strip was gently knocked to remove air bubbles and loaded into the Genie II machine (OptiGene Ltd., Horsham, West Sussex, UK; purchased from Amplex BioSystems). Tests were run at 65 °C for 20 min.
Collection and processing of BCs
Clinical samples were BCs submitted as part of routine patient care to the laboratory from the University Hospital Jena between April and August 2015. Blood samples collected in BD BACTEC Plus Aerobic/F and Lytic/10 Anaerobic/F bottles (BD Diagnostics, Heidelberg, Germany) were incubated on a BACTEC FX instrument (BD Diagnostics). Positive BCs were sampled aseptically, Gram-stained, and streaked onto Columbia sheep blood agar, chocolate agar, Drigalski lactose agar, and Schaedler agar (Oxoid, Thermo Fisher Scientific) for overnight incubation at 37 °C. In parallel, an aliquot was prospectively tested using the LAMP eazyplex® assays MRSA, GN, and GP according to the results of Gram staining. Only one positive BC bottle per patient was tested.
LAMP testing of BCs
25 μL of BC broth were mixed with 500 μL of RALF and boiled for 2 min. After centrifugation at 4000 rpm for 1 min, 25 μL of the supernatant were added to each tube of the eazyplex® test strip containing the lyophilized master mix. The strip was gently knocked to remove air bubbles and loaded into the Genie II machine. Tests were run at 65 °C for 20 min. Amplification was measured by real-time fluorescence detection using a DNA intercalating dye. Depending on the result of Gram staining, the MRSA, GP, and GN tests were used for Gram-positive cocci in clusters, Gram-positive cocci arranged in chains or as diplococci, and Gram-negative rods, respectively.
Conventional species identification and AST
Isolates sampled from positive BCs were identified by Vitek MS (bioMérieux, Nürtingen, Germany). Preliminary resistance patterns by disk diffusion assay were evaluated according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints. AST was performed using Vitek 2 and minimal inhibitory concentration (MIC) interpretation according to EUCAST criteria. The production of ESBLs was verified by the combination disk method using cefotaxime (CTX) vs. CTX+clavulanic acid (CV) and cefpodoxime (CPD) vs. CPD+CV. The LAMP results on CTX-M were verified for phenotypic ESBL strains using the Verigene BC-GN array (Nanosphere, Northbrook, IL, USA; purchased from Thermo Fisher Scientific, Wesel, Germany). For staphylococci, cefoxitin disk diffusion assay was performed to confirm beta-lactam resistance.
Results and discussion
Overview
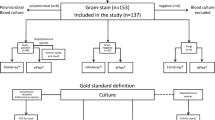
To investigate the potential of LAMP for BC diagnosis, three different assays were applied (Table 1). The eazyplex® MRSA assay is a commercial CE-labeled test. For the GP and GN in-house assays, verification was performed using reference strains selected from our collection of bacterial strains (Table 2). All enterococci and streptococci strains were correctly identified. For Gram-negatives, there were a small number of incorrectly identified Pseudomonas strains that showed signals for both Pseudomonas spp. and Enterobacteriaceae.
For the evaluation of clinical samples, a total of 370 positive BCs were investigated. The initial Gram staining was used to apply only the appropriate eazyplex® assay strip, thereby saving costs. The results of LAMP were compared with routine diagnosis species identification and AST. The sensitivity, specificity, and positive and negative predictive values of the eazyplex® assays for clinical BCs are summarized in Table 3. The eazyplex® LAMP tests demonstrated high sensitivity and specificity for the identification of species and resistance genes in BCs. One MRSA and two GN tests reported an invalid result (0.8%). Overall, 532/553 (96.2%) fluorescence signals of targets of the eazyplex® assays represented true-positive results, compared to routine species identification as the gold standard.
Staphylococci
A total of 140 BCs that showed growth of Gram-positive cocci in clusters were analyzed with the eazyplex® MRSA assay. Staphylococcus aureus was diagnosed with high accuracy (Table 3). Of the two specimens with a false-positive signal for S. aureus, S. epidermidis (1) and S. hominis (1) were grown. All cases of MRSA infections were correctly identified by the detection of mecA. No isolates with mecC could be found. Of the 78 S. epidermidis isolates that were identified phenotypically, six were not detected by eazyplex® (92.3% sensitivity). Twenty-nine BCs with Gram-positive cocci in clusters gave no species result in the eazyplex® assay but 17 of them were tested positive for mecA. Subcultures revealed CoNS other than S. epidermidis in 28 cases and Stomatococcus spp. in one case. All mecA signals for BCs with CoNS including S. epidermidis were concordant with the subsequent detection of oxacillin resistance. Two oxacillin-resistant strains that were found to be mecA- and mecC-negative were defined as false-negative results to calculate the sensitivity of mecA/mecC detection for the identification of beta-lactam resistance in CoNS in general (Table 3). One MRSA eazyplex® test reported an invalid result. Because S. hominis which was subcultured from the BC bottle is not covered by the targets of the assay, this result was excluded from the performance analysis.
Streptococci and enterococci
A total of 71 BCs were tested with the GP assay (Table 3). In 43 cases, Enterococcus spp. was detected. One false-positive result for E. faecalis came from a BC with growth of S. dysgalactiae ssp. equisimilis that also showed a Streptococcus spp. signal in the same test strip. eazyplex® assays that were positive for Enterococcus spp. but negative for E. faecalis revealed E. faecium (27), E. avium (2), and two false-positive test results for BCs that contained S. mitis and S. parasanguinis. The detection of vanA by the GP assay was concordant with phenotypic identification of vancomycin resistance for 13/13 E. faecium isolates (Table 3). No isolates were positive for vanB. Streptococci were identified in 20 BCs. There was only one false-negative test result for a BC that contained S. anginosus. True-positive eazyplex® test results revealed S. agalactiae (2), S. dysgalactiae spp. equisimilis (5), S. mitis (4), S. oralis (1), S. parasanguinis (3), S. sanguinis (3), and S. pneumoniae (2). Both cases of S. pneumoniae were also correctly identified by eazyplex® at the species level in the same test strip. One BC with a negative GP test result contained Peptostreptococcus asaccharolyticus. For seven further negative tests, the initial Gram stain was misinterpreted, resulting in the application of the GP test for BCs that contained CoNS.
Gram-negatives
A total of 159 BCs that showed growth of Gram-negative rods were analyzed with the GN assay (Table 3). Two false-positive results for E. coli were identified as K. oxytoca and A. junii by subculture diagnosis. Two false-positive results for K. pneumoniae were caused by cross-reactions of E. coli with the K. pneumoniae target. In both cases, E. coli was correctly identified in parallel. One false-negative K. pneumoniae sample only showed an Enterobacteriaceae signal. All E. coli and K. pneumoniae isolates that were tested positive for CTX-M genes by eazyplex® GN were subsequently confirmed as ESBL strains by phenotypic AST. Of the 19 E. coli ESBL isolates, two (10.5%) were CTX-M-negative and, therefore, could not be identified as ESBL by the GN assay. In Table 3, the CTX-M results are also evaluated in relation to the phenotypic identification of an ESBL strain in order to calculate the predictive values of the CTX-M target for identifying ESBL resistance. For K. pneumoniae, the concordance of ESBL resistance with detection of CTX-M was 100%. Only CTX-M beta-lactamases were considered as resistance markers in this study because of a very low incidence of carbapenemase-producing Enterobacteriaceae at the Jena University Hospital. It should be noted that other eazyplex® LAMP assays have also been successfully applied to detect carbapenemase genes in BCs that were spiked with carbapenem-resistant bacterial isolates [16]. There were 25 BCs that were true-positive for the Enterobacteriaceae family-level assay but showed no species-specific test result. Subculture identification revealed K. oxytoca (3), C. braakii (1), C. farmeri (1), C. freundii (1), C. koseri (2), E. aerogenes (1), E. cloacae (7), P. mirabilis (5), and S. marcescens (3). However, there were also 11 false-positive eazyplex® GN results for Enterobacteriaceae that were caused by BCs containing P. aeruginosa (6), M. osloensis (1), A. baumannii (1), A. junii (1), and H. influenzae (2). To cover a broad range of species of Enterobacteriaceae, ubiquitous primer targets of 16S rDNA must be chosen. Potential cross-reactions to other bacteria could not be fully excluded but false-positive signals typically occurred at significantly later threshold times than specific signals and specificity can be increased by reducing the running time cut-off (Table 4). When a cut-off of 15 min was defined, the specificity of the Enterobacteriaceae signal was increased to 91.3% (true-positive: 133, true-negative: 21, false-positive: 2, false-negative: 3). For two BCs that showed growth of P. mirabilis, the GN assay was invalid. All cases of P. aeruginosa infections were correctly identified as Pseudomonas spp. From nine BC bottles with a negative valid GN result, non-fermenters (5) and Bacteroides spp. (4) that are not covered by the eazyplex® primers were subcultured.
Time to result of LAMP for BC analysis
The threshold time for fluorescence intensity values ranged from 6.25 to 13.75 min (Table 4). The internal controls were detected between 7.5 and 9 min. With the exception of K. pneumoniae, false-positive signals were typically detected later than true-positive signals [mean values (SD): 15.25 (3.5) min, n = 21, vs. 8.5 (3.5) min, n = 532; p < 0.0001, unpaired t-test].
Mixed infections
From 15 BC bottles, mixed subcultures were obtained. Depending on the results of Gram staining, one or two eazyplex® tests were performed (Table 5). In 11 cases, eazyplex® assays produced correct results including the determination of resistance genes, compared to subculture identification (73%). It should be noted that, in two cases, S. epidermidis was not recognized because only the GP assay was applied based on the interpretation of the Gram stain. There were three cases for which the eazyplex® MRSA assay could not correctly differentiate mixed infections with staphylococci, including one false-positive test result for S. aureus.
Conclusions
The impact of rapid blood culture (BC) diagnostic tests on the health care of sepsis patients has been demonstrated in recent studies [17, 18]. The results of this study show that loop-mediated isothermal amplification (LAMP), as an alternative technique to polymerase chain reaction (PCR) assays, is a powerful tool for the rapid identification of common bacterial pathogens and their major resistance genes directly from positive BCs. LAMP is easy to handle and needs only a short running time for amplification when coupled to fluorescence real-time detection. There is no need for DNA purification because Bst polymerase tolerates serum and heparin [13, 14]. In principle, testing positive BCs with eazyplex® LAMP offers the possibility to generate results within half an hour, including Gram stain and sample preparation. Another advantage is that no expensive equipment is needed. Limitations of this study include the restricted pathogen panel of the assays. The eazyplex® strip format allows only a maximum of seven target genes and an inhibition control. The LAMP assays applied in this study only detect the most frequent sepsis pathogens and key resistance genes for staphylococci, enterococci, and Enterobacteriaceae. However, when the local hospital antibiogram is taken into consideration, these tests allow specific antibiotic treatment recommendations about one day earlier in comparison to classical BC diagnosis, thereby contributing to improved antibiotic stewardship for sepsis patients.
References
Zilberberg MD, Shorr AF, Micek ST, Vazquez-Guillamet C, Kollef MH (2014) Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit Care 18:596
Fitting C, Parlato M, Adib-Conquy M, Memain N, Philippart F, Misset B, Monchi M, Cavaillon JM, Adrie C (2012) DNAemia detection by multiplex PCR and biomarkers for infection in systemic inflammatory response syndrome patients. PLoS One 7, e38916
Vincent JL, Brealey D, Libert N, Abidi NE, O’Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M, Picard-Maureau M, Chalfin DB, Ecker DJ, Sampath R, Singer M; Rapid Diagnosis of Infections in the Critically Ill Team (2015) Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med 43:2283–2291
Rödel J, Karrasch M, Edel B, Stoll S, Bohnert J, Löffler B, Saupe A, Pfister W (2016) Antibiotic treatment algorithm development based on a microarray nucleic acid assay for rapid bacterial identification and resistance determination from positive blood cultures. Diagn Microbiol Infect Dis 84:252–257
Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ (2016) Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698
Rand KH, Delano JP (2014) Direct identification of bacteria in positive blood cultures: comparison of two rapid methods, FilmArray and mass spectrometry. Diagn Microbiol Infect Dis 79:293–297
Alby K, Daniels LM, Weber DJ, Miller MB (2013) Development of a treatment algorithm for streptococci and enterococci from positive blood cultures identified with the Verigene Gram-positive blood culture assay. J Clin Microbiol 51:3869–3871
Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD (2015) Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 34:487–496. doi:10.1007/s10096-014-2252-2
Southern TR, VanSchooneveld TC, Bannister DL, Brown TL, Crismon AS, Buss SN, Iwen PC, Fey PD (2015) Implementation and performance of the BioFire FilmArray® blood culture identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagn Microbiol Infect Dis 81:96–101
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, e63
Goldmeyer J, Li H, McCormac M, Cook S, Stratton C, Lemieux B, Kong H, Tang W, Tang YW (2008) Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol 46:1534–1536
Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD, Schrenzel J (2011) Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 62:41–48
García-Fernández S, Morosini MI, Marco F, Gijón D, Vergara A, Vila J, Ruiz-Garbajosa P, Cantón R (2015) Evaluation of the eazyplex® SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother 70:1047–1050
Findlay J, Hopkins KL, Meunier D, Woodford N (2015) Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother 70:1338–1342
Hinić V, Ziegler J, Straub C, Goldenberger D, Frei R (2015) Extended-spectrum β-lactamase (ESBL) detection directly from urine samples with the rapid isothermal amplification-based eazyplex® SuperBug CRE assay: proof of concept. J Microbiol Methods 119:203–205
Zboromyrska Y, Vergara A, Cosgaya C, Verger G, Mosqueda N, Almela M, Pitart C, Roca I, Marco F, Vila J (2015) Rapid detection of β-lactamases directly from positive blood cultures using a loop-mediated isothermal amplification (LAMP)-based assay. Int J Antimicrob Agents 46:355–356
Box MJ, Sullivan EL, Ortwine KN, Parmenter MA, Quigley MM, Aguilar-Higgins LM, MacIntosh CL, Goerke KF, Lim RA (2015) Outcomes of rapid identification for Gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy 35:269–276
Roshdy DG, Tran A, LeCroy N, Zeng D, Ou FS, Daniels LM, Weber DJ, Alby K, Miller MB (2015) Impact of a rapid microarray-based assay for identification of positive blood cultures for treatment optimization for patients with streptococcal and enterococcal bacteremia. J Clin Microbiol 53:1411–1414
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The study protocol for the evaluation of LAMP eazyplex® assays for clinical BCs was reviewed and approved by the ethics committee of the Friedrich Schiller University of Jena (4400-04/15).
Funding information
This study was supported by internal funding from the Jena University Hospital and by a grant from the German Federal Ministry of Education and Research (BMBF, 13N13890).
Conflict of interest
J. Rödel received financial support from Amplex BioSystems to attend the 67th Annual Meeting of the German Society for Hygiene and Microbiology in 2015. No funding was received from the manufacturer of eazyplex® tests for the purpose of the study.
Rights and permissions
About this article
Cite this article
Rödel, J., Bohnert, J.A., Stoll, S. et al. Evaluation of loop-mediated isothermal amplification for the rapid identification of bacteria and resistance determinants in positive blood cultures. Eur J Clin Microbiol Infect Dis 36, 1033–1040 (2017). https://doi.org/10.1007/s10096-016-2888-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2888-1