Abstract
The workup and interpretation of urine cultures is not always clear-cut, especially for midstream samples contaminated with commensals. Standard urine culture (SUC) protocols are designed in favor of growth of uropathogens at the expense of commensals. In selected clinical situations, however, it is essential to trace fastidious or new uropathogens by expanding the urine culture conditions (EUC). The aim of our study was to map the microflora in midstream urine specimens from healthy controls by means of EUC, in view of the interpretation of bacterial culture results in symptomatic patients. Midstream urine specimens from 101 healthy controls (86 females and 15 males) were examined using both SUC and EUC. Whilst 73 % of samples examined by SUC showed no growth at 103 colony-forming units (CFU)/mL, 91 % of samples examined by EUC grew bacterial species in large numbers (≥104 CFU/mL). Asymptomatic bacteriuria, as defined by the European guidelines for urinalysis, was detected in six samples with both protocols. EUC revealed 98 different species, mostly Lactobacillus, Staphylococcus, Streptococcus, and Corynebacterium. None of the samples grew Staphylococcus saprophyticus, Corynebacterium urealyticum, or Aerococcus urinae. Samples from females contained higher bacterial loads and showed higher bacterial diversity compared to males. Midstream urine of healthy controls contains large communities of living bacteria that comprise a resident microflora, only revealed by EUC. Hence, the use of EUC instead of SUC in a routine setting would result in more sensitive but less specific results, requiring critical interpretation. In our view, EUC should be reserved for limited indications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Urinary tract infections (UTI) are common and urine cultures constitute an important workload in the microbiology lab. The workup and interpretation of urine cultures is not always clear-cut, especially when non-invasive sampling techniques (i.e., voided midstream) are used, contaminating the samples with microflora of the skin, perineum, urethra, or vagina. For urine culture, standard media such as blood agar plate (BAP), MacConkey agar (MAC), or cystine lactose electrolyte deficient (CLED) agar are recommended and an incubation for longer than 18 to 24 h is usually not advised [1, 2]. In selected clinical situations, more controversial fastidious uropathogens (like Gardnerella vaginalis) and new uropathogens (like Aerococcus spp., Corynebacterium urealyticum, and Actinobaculum schaalii) should be traced, using prolonged incubation of BAP [3]. Expanded urine culture conditions (EUC), such as larger volumes of urine, extended incubation times, and incubation in varied atmospheric conditions, reveal many species, mostly commensals. The aim of this study was to map the microflora in voided midstream urine specimens from healthy controls by means of EUC, in view of the interpretation of bacterial culture results in symptomatic patients.
Materials and methods
Following approval by the local Ethical Committee, 101 healthy volunteers (86 female and 15 male, aged between 23 and 65 years) gave written consent for the collection and analysis of their urine for research purposes. During the recruitment (August–October 2015), the following exclusion criteria were taken into account: abnormal vaginal discharge; vaginal pruritus; dysuria; history of cystitis in the past three months; history of at least two cystitis episodes in the past year; use of antibiotics in the last month; recent gynecological, nephrological, or urological disease; pregnancy; diabetes, or other chronic disease. From each individual, a clean-voided midstream urine was collected in a sterile receptacle and transported to the laboratory at room temperature within two hours after collection. On arrival, a Uricult dipslide with CLED and MAC (Mediphos, Renkum, The Netherlands) was immersed and incubated in ambient atmosphere at 35 °C for 24 h (standard urine culture, SUC). Next, a Trypticase soy agar with 5 % sheep blood and a Schaedler agar (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) were inoculated with 10 μL of urine and incubated in ambient atmosphere and anaerobically, respectively, at 35 °C for seven days (EUC). Finally, 2 mL of urine was aspirated in a Urine Monovette tube (Sarstedt, Nümbrecht, Germany) for urinalysis on the UF-1000i apparatus (Sysmex Corporation, Kobe, Japan). When interpreting the results of urinalysis, pyuria was considered significant in case of more than 25 leukocytes per μL of urine in the absence of over 25 epithelial cells per μL of urine. For SUC, the colony count density on the agar surfaces of the dipslides was compared with the colony density chart provided to obtain a semiquantitative colony count in colony-forming units (CFU)/mL of urine. For EUC, a consensus colony count (CFU/mL) was made of the two agars and all CFUs with distinct morphological appearance were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the MALDI Biotyper 3.0 real-time classification software (Bruker Daltonics, Billerica, MA, USA). A single measurement was performed once for each culture isolate. A score between 2000 and 3000 allowed for species-level identification, a score between 1700 and 1999 for genus-level identification, and a score below 1700 was unreliable. The European guidelines for urinalysis [4] were used to count the number of primary, secondary, and doubtful uropathogens, as well as the cases of asymptomatic bacteriuria.
Results
The majority of the samples [74/101 (73.3 %)] showed growth with a total colony count of less than 103 CFU/mL by SUC. In contrast, not a single sample examined by EUC showed growth with a total colony count of less than 103 CFU/mL, and nearly all samples grew bacterial species in numbers ≥104 CFU/mL: [92/101 (91.1 %), Fig. 1]. When comparing female (n = 86) and male (n = 15) individuals, midstream samples from females contained higher bacterial loads: the total colony count exceeded 104 CFU/mL in 97.7 % of females versus 53.3 % of males.
Only 10/101 samples (9.9 %) had significant pyuria. Asymptomatic bacteriuria was detected in six samples with both SUC and EUC: three samples of females with ≥105 CFU/mL Escherichia coli, one sample of a male with ≥105 CFU/mL Enterococcus faecalis, one sample of a female with ≥105 CFU/mL Staphylococcus epidermidis, and one sample of a female with ≥105 CFU/mL of both Escherichia coli and Enterococcus faecalis. Only the latter sample showed significant pyuria.
Overall, 539 isolates were recovered by EUC, belonging to 41 different genera and 98 different species. The bacterial diversity was higher in females, reflected by the average recovery of six different species in samples from females versus three different species in samples from males (Fig. 2). The complete results of EUC are available as Supplemental Material Tables 1 (females) and 2 (males). Overall, the top five prevalent genera were Lactobacillus (n = 101), Staphylococcus (n = 88), Streptococcus (n = 57), Corynebacterium (n = 53), and Propionibacterium (n = 24). The five most prevalent species were Staphylococcus epidermidis (n = 41), Streptococcus anginosus (n = 36), Lactobacillus jensenii (n = 35), Lactobacillus crispatus (n = 30), and Gardnerella vaginalis (n = 20). If present, Lactobacillus spp. and Gardnerella vaginalis were recovered in the largest numbers (Fig. 3). We cultured 1.7 % (9/539) primary uropathogens (nine Escherichia coli), 3.2 % (17/539) secondary uropathogens (11 Enterococcus faecalis, two Klebsiella pneumoniae, two Proteus mirabilis, one Staphylococcus aureus, one Citrobacter koseri), and 18.9 % (102/539) doubtful uropathogens (41 Staphylococcus epidermidis, 20 Staphylococcus haemolyticus, 13 Streptococcus agalactiae, 11 Staphylococcus hominis, six Staphylococcus lugdunensis, three Staphylococcus capitis, two Staphylococcus simulans, two Staphylococcus spp., one Acinetobacter lwoffii, one Staphylococcus caprae, one Staphylococcus saccharolyticus, one Staphylococcus warneri). None of the samples grew Staphylococcus saprophyticus, Corynebacterium urealyticum, or Aerococcus urinae. Actinobaculum schaalii, a novel uropathogen, was isolated in one female and one male individual in small numbers (≤103 CFU/mL) and only once with significant pyuria. Alloscardovia omnicolens, a recently suggested uropathogen [5], was recovered from 13 females in varying numbers (≥103 CFU/mL) and only once with significant pyuria.
Discussion
Many studies describe the existence of communities of living bacteria that comprise a urinary resident microflora [6–15]. Besides this microflora, voided urine samples mostly contain vulvovaginal or urethral bacteria. In our study, the majority (73.3 %) of the midstream samples showed growth with a total colony count of less than 103 CFU/mL by SUC, which equals the yield reported by others in urine collected via transurethral catheter (71.1–73.8 %) [11, 14]. It should be emphasized that every case of asymptomatic bacteriuria showed growth with a total colony count of ≥105 CFU/mL by SUC.
By culturing midstream urine of healthy controls under expanded conditions, we confirmed that the urinary microbiota is ubiquitous, irrespective of symptomatology, as growth was observed in every single sample. In contrast, reports on EUC using urine collected via transurethral catheter showed growth in only 80–96 % of cases [14, 15]. As we demonstrated that all midstream urine samples contain bacterial species in large numbers, the authors want to warn against equaling the presence of bacteria detected by urinary flow analyzers to the diagnosis of UTI, leading to wrongful use of antibiotics.
Identification of all species grown by EUC shows that the urinary microbiota consists of numerous species. In our study, we identified 41 different genera and 98 different species, reflecting a greater diversity than reports on EUC of urine collected via transurethral catheter (28–35 different genera and 75–85 different species) [14, 15]. However, the abundance of Lactobacillus spp., Staphylococcus spp., and Streptococcus spp. in our study corroborates the findings of others [8, 14, 15]. Let it be noted that, as we collected midstream urine samples, our study does not allow to differentiate between urinary resident microflora in the bladder or urinary tract and vulvovaginal or urethral contamination.
SUC protocols are designed to quickly detect a selected group of known uropathogens, but hamper the growth of the vast majority of other bacteria that require special nutrients, grow slowly, cannot tolerate oxygen, or are present in small numbers [16]. Quantitative and qualitative criteria have been developed for the interpretation of SUC results, allowing to report some urogenital bacteria as potentially significant if they are present in amounts ten-fold more than other microbiota [2]. Analysis of our findings learns that this approach is rather arbitrarily, as commensals are present in large numbers in almost all voided specimens, with the detection and density depending on the strains present and culture conditions, i.e., richness of media and duration of incubation. For instance, vaginal lactobacilli can appear after one day of incubation, but some isolates appear only after several days or only if incubated anaerobically. Some authors suggest that EUC on invasive samples should be considered in selected clinical situations, i.e., as a supplemental test when urine samples of individuals with UTI-like symptoms show no growth by SUC and for use on urine of individuals with recurrent UTI [15]. In casu, fastidious bacterial species as Aerococcus spp., Corynebacterium urealyticum, Actinobaculum schaalii, or Gardnerella vaginalis, or other new or previously unappreciated uropathogens which can only be detected by means of EUC, could be designated as the causal agent of the UTI. Given the ubiquity of commensals yielded by EUC, we argue in favor of more profound research into the growth of fastidious bacteria in order to further specify the interpretative criteria, thereby avoiding misdiagnosis.
In general, contamination of midstream voided urine is considered uncommon in males and low bacterial counts with uropathogens are more often considered clinically meaningful [17]. Our data, however, demonstrate that every single midstream urine sample is contaminated with commensal bacteria, irrespective of the sex. These findings are corroborated by others, who mapped the urinary microbiome of men [18].
Our study has some limitations. When reading the incubated agar media, all species with different colony morphology were identified. As different species may have similar morphological appearance, we possibly underestimated the diversity of the urinary microbiota. This diversity could also be influenced by some isolates that were not fully identifiable by MALDI-TOF MS and were reported at the genus level. Besides, we only included asymptomatic individuals and cannot speak about the possible differences in the urinary microbiota of healthy versus symptomatic individuals. Also, our study design does not allow differentiation between urinary resident microflora in the bladder or urinary tract and vulvovaginal or urethral contamination.
Conclusion
To our knowledge, our study is the first to document the presence of live bacterial species in voided midstream samples by means of expanded urine culture conditions (EUC). The use of EUC instead of standard urine culture (SUC) in a routine setting would result in more sensitive but less specific results, as the urinary microbiota is present in healthy controls as well. Our study exposes the need for extensive evaluation of the clinical value of EUC in the diagnostic workup of urinary tract infections (UTI). Because the results of EUC require critical interpretation and SUC seems to be equivalent to EUC when it comes to the detection of asymptomatic bacteriuria, in our view, EUC should be reserved for limited indications.
References
Wilson ML, Gaido L (2004) Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis 38(8):1150–1158
Garcia LS (2010) Clinical microbiology procedures handbook, 3rd edn. ASM Press, Washington DC, pp 410–440
McCarter YS, Burd EM, Hall GS, Zervos M (2009) Cumitech 2C: laboratory diagnosis of urinary tract infections (Cumitechs). ASM Press, Washington DC
Aspevall O, Hallander H, Gant V, Kouri T (2001) European guidelines for urinalysis: a collaborative document produced by European clinical microbiologists and clinical chemists under ECLM in collaboration with ESCMID. Clin Microbiol Infect 7(4):173–178
Brown MK, Forbes BA, Stitley K, Doern CD (2016) Defining the clinical significance of Alloscardovia omnincolens in the urinary tract. J Clin Microbiol 54(6):1552–1556
Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK, Torralba M, Nelson KE, Groah SL (2012) Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 10:174
Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, Drake MJ (2013) The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol 3:41
Siddiqui H, Nederbragt AJ, Lagesen K, Jeansson SL, Jakobsen KS (2011) Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol 11:244
Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L (2012) Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 50(4):1376–1383
Dong Q, Nelson DE, Toh E, Diao L, Gao X, Fortenberry JD, Van der Pol B (2011) The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS One 6(5):e19709
Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, Kliethermes S, Schreckenberger PC, Brubaker L, Gai X, Wolfe AJ (2014) The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio 5(4):e01283-14
Nienhouse V, Gao X, Dong Q, Nelson DE, Toh E, McKinley K, Schreckenberger P, Shibata N, Fok CS, Mueller ER, Brubaker L, Wolfe AJ, Radek KA (2014) Interplay between bladder microbiota and urinary antimicrobial peptides: mechanisms for human urinary tract infection risk and symptom severity. PLoS One 9(12):e114185
Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, Malone-Lee J (2013) Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol 51(7):2054–2062
Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC (2014) Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 52(3):871–876
Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, Brubaker L, Wolfe AJ, Mueller ER, Schreckenberger PC (2016) The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol 54(5):1216–1222
Kass EH (1962) Pyelonephritis and bacteriuria. A major problem in preventive medicine. Ann Intern Med 56:46–53
Franz M, Hörl WH (1999) Common errors in diagnosis and management of urinary tract infection. I: pathophysiology and diagnostic techniques. Nephrol Dial Transplant 14(11):2746–2753
Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, Sodergren E, Weinstock GM, Diao L, Fortenberry JD (2010) Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One 5(11):e14116
Acknowledgments
The authors would like to thank the healthy volunteers for their cooperation and the technicians of the Department of Laboratory Medicine for their technical contributions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received.
Conflict of interest
The authors have no conflict of interest.
Ethical approval
The study was approved by the Ethical Committee of the Ghent University Hospital with Belgian registration number B670201525210.
Informed consent
All included healthy volunteers gave written informed consent.
Rights and permissions
About this article
Cite this article
Coorevits, L., Heytens, S., Boelens, J. et al. The resident microflora of voided midstream urine of healthy controls: standard versus expanded urine culture protocols. Eur J Clin Microbiol Infect Dis 36, 635–639 (2017). https://doi.org/10.1007/s10096-016-2839-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2839-x


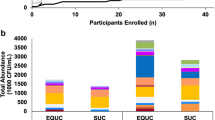


 ) and EUC (■) of midstream urine samples of 85 female (a) and 15 male healthy volunteers (b)
) and EUC (■) of midstream urine samples of 85 female (a) and 15 male healthy volunteers (b)
 ) and females (■)
) and females (■)
 ≥103 to <104 CFU/mL;
≥103 to <104 CFU/mL;  ≥104 to <105 CFU/mL; ■ ≥105 CFU/mL)
≥104 to <105 CFU/mL; ■ ≥105 CFU/mL)