Abstract
Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae have been described worldwide, but there are few reports on the carriage of these bacteria in Cameroon. In order to investigate the types of ESBLs and to analyse some risk factors associated with ESBL carriage, faecal samples were collected between 3 January and 3 April 2009 from hospitalised patients at Yaounde Central Hospital and at two hospitals in Ngaoundere, Cameroon. Enterobacterial isolates resistant to third-generation cephalosporins were screened for ESBL production using the double-disk synergy test. Polymerase chain reaction (PCR) and DNA sequencing were performed in order to find out the different types of ESBL genes in presumptive ESBL-positive isolates. During the study period, a total of 121 different patients were screened for ESBL carriage. The prevalence among these patients whose faecal samples were found to contain ESBL-producers was 55.3 % (67/121). According to a univariate analysis, hospitalisation during the previous year was found to be associated with ESBL carriage. Of the 71 bacteria isolated, Escherichia coli was predominant and represented 48 % of all isolates. ESBL characterisation revealed two types of ESBLs, CTX-M-15 (96 %) and SHV-12 (4 %). The present study emphasises the importance of screening for ESBLs in laboratories in African countries. The monitoring and detection of ESBL-producing bacteria are important in the setting up of appropriate treatment of patients and to ensure effective infection control efforts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae are mostly encoded by plasmids, although some of them are from chromosomal origin [1–3]. ESBLs are able to hydrolyse a wide range of β-lactam antibiotics, including second- and third-generation cephalosporins [4]. The first ESBLs described were derived from the two “classical” β-lactamase families—TEM and SHV—from which mutations have since occurred. In the last few years, however, new families of ESBL enzymes have been discovered. CTX-M-type ESBLs, a group of enzymes with an increased activity against cefotaxime, have become widespread in Europe and other continents [3, 5]. Since their first description in 1983 in Germany [6], ESBLs have been described worldwide, but in African countries, data are still very scarce. In Cameroon, although rare, studies have reported the presence of ESBL types SHV and CTX-M among inpatients in the city of Yaounde [7–9]. Moreover, although in Yaounde some data on ESBLs are available, in other cities in Cameroon, there is no information on antibacterial resistance. So, to increase the database on the subject, we thought it would be interesting to study and determine the prevalence of ESBL-producing Enterobacteriaceae isolated from the faecal flora of inpatients more than ten years after the first study [9] at the same hospital (Yaounde Central Hospital) and in two other Cameroonian hospitals located in the Adamawa province (city of Ngaoundere, located 858 km from Yaounde).
Materials and methods
Study setting
During a period of three months (from 3 January 2009 to 3 April 2009), faecal samples were collected from hospitalised patients for more or less 48 h in three different hospitals in Cameroon. Written informed consent was obtained from each patient who volunteered to participate in the study. All participants were asked to fill out a standardised questionnaire, including general demographics data, diagnosis at admission, hospital stays in the previous year and antibiotic use in the past three months. The three participating hospitals were defined as follows:
-
1.
Yaounde Central Hospital is the largest medical institution located in the city of Yaounde (political capital of Cameroon) in the central province, covering a population of about 3,525,664 inhabitants. This hospital has 650 beds, annually admits more than 20,000 inpatients and is comprised of 20 wards divided into five unit specialisations (medicine and specialities; surgery; intensive care unit; emergency; gynaecology; obstetrics and maintenance unit). The recruitment of patients was done in the medical unit and specialities (in cardiology/diabetes and gastroenteritis wards), emergency and intensive care units.
-
2.
Ngaoundere Protestant Hospital and (3) Ngaoundere Regional Hospital are the two largest hospitals located in the city of Ngaoundere (in the Adamawa province, covering a population of about 1,015,622 inhabitants). These hospitals are intermediate-level hospitals in the health system in Cameroon, with about 150 beds each. The annual number of hospitalisations ranges from 5,000 to 10,000 patients across many wards (medicine, surgery, medical imaging, maternity, paediatrics, emergency, intensive care unit, operating room and maintenance unit). In these two hospitals, patients were recruited in the medicine, surgery, intensive care unit and emergency wards.
Detection of ESBL-producing isolates in faecal samples
A total of 0.5 g of each faecal sample was suspended in 5 mL of sterile saline, and aliquots of 50 μl were streaked onto two selective media, Drigalski and MacConkey agars, supplemented respectively with cefotaxime (1.5 mg/L) and ceftazidime (2 mg/L), enabling the detection of Gram-negative bacteria resistant to these antibiotics. Plates were incubated for 24 h at 35 ± 2 °C. One colony representing each distinct colonial morphotype was subcultured on MacConkey agar plates and was analysed further. All isolates that showed growth were screened for ESBL production by using both the resistance phenotype and the combined double-disk synergy test with 30 μg amoxicillin/clavulanate (20 μg of amoxicillin; 10 μg of clavulanic acid), 30 μg cefotaxime, 30 μg ceftazidime and 30 μg cefepime disks (BBL, USA), which were applied at a 30 mm distance from amoxicillin/clavulanate [10].
Identification and antimicrobial susceptibility testing
Only the isolates with a positive synergy test—the “presumptive ESBL-producers”—were identified by mass spectrometry using the BioTyper MALDI-TOF mass spectrometer (Bruker). For such ESBL-positive isolates, the minimal inhibitory concentration (MIC) to the following drugs was determined with the Vitek 2 system (bioMérieux, France): temocillin, meropenem, amikacin, gentamicin, ciprofloxacin, nitrofurantoin and trimethoprim/sulphamethoxazole. The results enabled us to define the isolates as being susceptible or resistant according to the criteria recommended by the Clinical and Laboratory Standards Institute (CLSI) [11]. All the presumptive ESBL-producers were further analysed by polymerase chain reaction (PCR) aimed at detecting ESBL genes (one isolate of each morphotype from each patient).
Automated DNA isolation
Bacteria were grown overnight on MacConkey agar plates at 35 ± 2 °C. Subsequently, colonies were picked up using a sterile 1-μl plastic loop and transferred into 400 μl sterile water. Total DNA was extracted with the Maxwell automat (Promega, USA) according to the manufacturer’s protocol. DNA was eluted in a final volume of 200 μl. The extracted DNA was stored at −70 °C for further analysis.
PCR detection and sequencing of ESBLs
All isolates were initially screened for the presence of bla TEM-, bla SHV- and bla OXA-like genes using a multiplex PCR protocol previously described by Dallenne et al. [12]. PCR amplification of bla CTX-M-1 group alleles was carried out with primers CTX-M3G-F and CTX-M3G-R, as described previously using a simplex PCR according to a previously published method [13]. The primers used for PCR amplification are presented in Table 1. Specific PCR amplification and DNA sequencing of the PCR products were used to determine whether the bla TEM, bla SHV, bla OXA and bla CTX-M-1 group alleles were present and to characterise the type of β-lactamase belonging to each family. PCR products (multiplex and simplex PCR) were visualised after electrophoresis at 100 V for 1 h on a 2 % agarose gel containing ethidium bromide. A 100-bp DNA ladder (Promega, USA) was used as a marker size. PCR products were purified using the Wizard® kit (Promega, USA) and sequenced using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequence alignment and analyses were performed online using the BLAST program available on the National Center for Biotechnology Information web site (http://blast.ncbi.nlm.nih.gov/).
Statistical analysis
Statistical analysis was performed with Epi Info version 3.5.3 [Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA]. Pearson’s chi-square analysis or Fisher’s exact test, when appropriate, was used for the univariate comparison of all variables. Associations between ESBL carriage and the variables studied were determined by calculation of the odds ratios (ORs) along with 95 % confidence intervals (CIs). A value of p < 0.05 was considered to be statistically significant.
Results
Patient characteristics
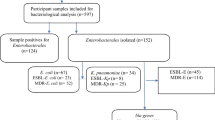
During the study period, a total of 121 patients were screened for ESBL production: 56 patients were from Yaounde Central Hospital (cardiology/diabetes, 28 patients; gastroenteritis, 26; emergency, 1; and intensive care unit, 1) and 65 patients were from the two Ngaoundere hospitals (medicine, 41 patients; surgery, 20; emergency, 2; and intensive care unit, 2). Overall, for these patients, the mean age was 46.81 ± 19.86 years and males accounted for 57 % of the study participants. The diagnosis at admission is presented in Table 2. Of all the diseases recorded, diarrhoea and diabetes mellitus were the most prevalent, representing 27.2 and 22.3 %, respectively. Before inclusion in the study, 26 patients had been admitted to hospital during the previous year and 49 patients had received antibiotics during the last 3 months. The names of the types of antibiotics used are also reported in Table 2.
Prevalence of ESBL-producers
Of the 121 cultures tested, 67 grew on both of the selective media and were designated presumptive ESBL-producers. All presumptive ESBL-producing Enterobacteriaceae were confirmed as ESBL-producers by PCR.
Overall, the prevalence of ESBL-producers among inpatients was 55.3 % (67/121). Among those patients found to be carrying ESBL-producers, ten were carriers within 48 h of admission to hospital and were, therefore, probably already colonised before admission. The other 57 carriers had already been hospitalised for more than 48 h when screening was done. Of the 67 positive cultures tested, 71 bacteria were isolated (we found one patient colonised with three isolates and two patients colonised with two isolates at Yaounde Central Hospital). The prevalence of ESBL-producers at Yaounde Central Hospital was significantly higher than the prevalence of ESBL-producers in the two Ngaoundere hospitals [73.2 % (41/56) and 40 % (26/65), respectively, p < 0.000] (see Table 2). According to the wards where patients were hospitalised, the prevalence of ESBL-producers at the Yaounde Central Hospital was 67.8 % (19/28) in cardiology/diabetes, 76.9 % (20/26) in gastroenteritis and 100 % (1/1) in the remaining services (emergency and intensive care unit). Similarly, in the two Ngaoundere hospitals, the prevalence of ESBL-producers was 39 % (16/41) in medicine, 40 % (8/20) in surgery and 50 % (1/2) in the remaining services (emergency and intensive care unit).
Potential associated factors for ESBL carriage
As shown in Table 2, when comparing ESBL-producers and non-ESBL-producers among the 121 hospitalised patients, hospitalisation during the previous year appeared to be associated with ESBL carriage (p < 0.000). However, no significant association was found between ESBL carriage and wards where the patient was hospitalised.
Bacterial identification and susceptibility
Of the 71 bacteria isolated, 34 (47.8 %) were Escherichia coli, 28 (39.4 %) Klebsiella spp. (27 K. pneumoniae and 1 K. oxytoca) and 9 (12.6 %) Enterobacter cloacae. ESBL-producing E. coli was the species frequently isolated in the two Ngaoundere hospitals (61.5 %), whereas at Yaounde Central Hospital, Klebsiella spp. were predominant (51.1 %).
The susceptibility profile of strains from the different hospitals to the antibiotics tested was relatively similar. All isolates were susceptible to temocillin, meropenem and amikacin.
Regardless of the hospital, E. cloacae was resistant to gentamicin (100 %), ciprofloxacin (90 %), trimethoprim/sulphamethoxazole (100 %) and nitrofurantoin (90 %).
Klebsiella spp. showed resistance to gentamicin (74.5 %), trimethoprim/sulphamethoxazole (100 %) and nitrofurantoin (67.8 %) but a good susceptibility to ciprofloxacin (60.5 %).
E. coli showed resistance to gentamicin (85 %), ciprofloxacin (93.7 %) and trimethoprim/sulphamethoxazole (100 %). However, the susceptibility of strains to nitrofurantoin was different between the hospital sites. Strains from Yaounde Central Hospital showed a good susceptibility (77.8 %) compared to strains from the two Ngaoundere hospitals (43.8 %).
β-lactamase characterisation
Patients shared the same ESBL types regardless of their hospital, and ESBL characterisation revealed two types of ESBLs: CTX-M-15 (96 %) and SHV-12 (4 %). The CTX-M-15 gene was always associated with other β-lactamase genes: OXA-1 and TEM-1 (34 %); TEM-1 alone (22 %); OXA-1 alone (19 %); TEM-1 and SHV-1 (13 %); TEM-1, OXA-1 and SHV-1(10 %); OXA-1 and SHV-1 (2 %) (Table 3).
Discussion
The present study was conducted to evaluate the prevalence of ESBL-producing Enterobacteriaceae among inpatients in three Cameroonian hospitals. Our results showed high levels of gut colonisation by ESBL-producing Enterobacteriaceae among inpatients (55.3 %), much higher than that observed in hospitals in other countries: Philippines (29.9 %) [14], Croatia (9.9 %) [15], Iran (16.8 %) [16], Portugal (39 %) [17], Indonesia (21 %) [18] and in some hospitals in low-income countries: Benin (22 %) [19], Tanzania (28.2 %) [20] and Ethiopia (15 %) [21].
The high prevalence of ESBL-producing Enterobacteriaceae described here was also due to the number of asymptomatic carriers entering hospitals. Indeed, in this study, 10 (14.9 %) patients were carriers of ESBL-producing Enterobacteriaceae within 48 h of admission to hospital and were, therefore, probably colonised before their admission, while for the other 57 carriers, the source of ESBL infection was unknown. Some studies have demonstrated the role of community transmission of ESBL-producing Enterobacteriaceae. Mirelis et al. observed a significant increase in the frequency of ESBL carriers in faecal samples from outpatients attending hospital. These patients came to hospital from the community, carrying strains that expressed ESBL [22]. Moreover, the accumulation of carriers in a catchment population links all of the institutions that serve it; carriers may shed antibiotic-resistant bacteria (ARB) for years but remain undetected, transmitting ARB to others as they move between hospitals, long-term care facilities and the community [23]. Recently, a publication by Lonchel et al. found a prevalence of ESBL-producers of approximately 23.1 % among outpatients (who visited the bacteriology laboratory of Ngaoundere Protestant Hospital with a stool sample requested by their general practitioner) and a prevalence of 6.7 % in faecal samples from student volunteers (the student volunteers at the University of Ngaoundere were attending the clinic for their annual medical examination) [24].
In the present study, the prevalence of ESBL-producers at Yaounde Central Hospital was significantly higher than the prevalence obtained at the two Ngaoundere hospitals (p < 0.000). Moreover, an earlier study conducted between 1995 and 1998 at Yaounde Central Hospital found a prevalence of about 12 % of ESBL-producers [9]. In 2009, in the same hospital, we found a dramatic increase in the prevalence of ESBL-producing Enterobacteriaceae to around 72.3 % (personal communication) from two wards of this hospital (cardiology/diabetes and gastroenteritis). Our results are likely to reflect the nosocomial spread and dissemination of ESBL-producing Enterobacteriaceae within the hospital. In the absence of infection control measures, ESBL-producing Enterobacteriaceae could readily pass horizontally from patient to patient. Due to a number of limitations, we were not able to determine the relatedness between isolates recovered from different patients.
In the two Ngaoundere hospitals, the prevalence of ESBL-producing Enterobacteriaceae found was also high: 44 % in Ngaoundere Protestant Hospital and 30 % in Ngaoundere Regional Hospital. To our knowledge, the present study is the first survey in the area and underlines the great need to apply proper empiric therapy through the implementation of methods of screening for ESBL-producing bacteria, which are currently lacking in laboratories in Cameroon.
We also investigated the potential associated factors for ESBL carriage and found no association between ESBL carriage and wards where the patient was hospitalised. However, hospitalisation during the previous year was found to be associated with ESBL carriage. This factor has been widely reported as a major cause of the development of infection by ESBL-producing Enterobacteriaceae in many studies [25–28].
In our study, we should sound the alarm regarding antibacterial resistance. Indeed, most of the patients were admitted with a history of antibiotic consumption. Patients had used an antibiotic or a combination of several antibiotics for empiric treatment. Some antibiotics were prescribed by medical professionals, while others were not. In general, there are no restrictions to the use of antibiotics in Cameroon or in many African countries, leading to a greater consumption of broad-spectrum antibiotics by the population. Prior antibiotic use has previously been described in many studies as a risk factor associated with ESBL carriage [25, 28–30].
Among the collected isolates, E. coli and K. pneumoniae were the main species identified. ESBL-producing E. coli and K. pneumoniae remain the predominant organisms harbouring ESBL worldwide [5]. The majority of our ESBL-producing Enterobacteriaceae was multiresistant; specifically, most were resistant to third-generation cephalosporins, ciprofloxacin, gentamicin and trimethoprim/sulphamethoxazole. These results are in line with the findings of previous studies in other African countries [16, 19, 31]. However, we found that drugs such as amoxicillin, gentamicin, trimethoprim/sulphamethoxazole and ceftriaxone were often used by patients in our study. These antibiotics are widely prescribed in Cameroon because of their widespread availability and low cost on the market. The previous use of fluoroquinolone (ciprofloxacin) appeared to have been an associated factor for ESBL carriage (p < 0.005) [24]. The reduction of efficacy of these drugs in the treatment of infections caused by ESBL-producing Enterobacteriaceae has been shown in several studies [16, 19, 21, 26, 32, 33] and their use should, therefore, be limited.
The results of PCR and DNA sequencing of ESBL genes found that all of the cefotaxime- and ceftazidime-resistant isolates had the CTX-M-15 enzyme. CTX-M enzymes (especially CTX-M-15) have emerged worldwide and have been identified in European, American and Asian countries [17, 34–37]. In African countries, the CTX-M-15 gene has been found in Tanzania [20, 38], South Africa [34, 39], Algeria [40–42], Nigeria [43], Egypt [31], the Central African Republic [44] and Tunisia [45]. The gene has also been found in Hungary [46] and Colombia [47]. In our study, the CTX-M-15 gene was found to be always associated with other β-lactamase genes (TEM-1, OXA-1 and SHV-1). The CTX-M-15 gene was found in 96 % of the isolates. Our results are similar to those regarding the distribution of the CTX-M-15 ESBL in Enterobacteriaceae in studies performed in other countries: Indonesia, 94.5 % of isolates [18]; Tunisia, 91 % [45]; Egypt, 89 % [31]; Mali, 83 % [48] and Algeria, 81 % [49]. It is clear that CTX-M-15 ESBLs are becoming widely distributed throughout Africa.
The other reported ESBL gene in the present study was SHV-12. This gene was found in K. pneumoniae and E. cloacae. The CTX-M-15 ESBL and SHV-12 ESBL were previously described in Cameroon [9, 24, 50] and, recently, types SHV-5 and -27 were also found [7]. As far as other African countries are concerned, type CTX-M-12 has been found in Kenya [51], CTX-M-14, -15 and -27 in Egypt [52] and CTX-M-3 and -15 in the Central African Republic [44].
Our results highlight a high prevalence of the faecal carriage of ESBL-producing Enterobacteriaceae in Cameroonian hospitals and denote the role of the community as a reservoir for the dissemination of ESBLs within hospitals. The present study emphasises the importance of screening for ESBLs in the laboratory. The monitoring and detection of ESBL-producing bacteria are important to ensure the appropriate treatment of patients and effective infection control efforts.
References
Song W, Kim J, Bae IK, Jeong SH, Seo YH, Shin JH, Jang SJ, Uh Y, Shin JH, Lee MK, Lee K (2011) Chromosome-encoded AmpC and CTX-M extended-spectrum beta-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob Agents Chemother 55(4):1414–1419
Fabre L, Delaune A, Espie E, Nygard K, Pardos M, Polomack L, Guesnier F, Galimand M, Lassen J, Weill FX (2009) Chromosomal integration of the extended-spectrum beta-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob Agents Chemother 53(5):1808–1816
Cantón R, González-Alba JM, Galán JC (2012) CTX-M enzymes: origin and diffusion. Front Microbiol 3:110
Rahal JJ (2000) Extended-spectrum beta-lactamases: how big is the problem? Clin Microbiol Infect 6(Suppl 2):2–6
Colodner R, Raz R (2005) Extended-spectrum beta-lactamases: the end of cephalosporins? Isr Med Assoc J 7(5):336–338
Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11(6):315–317
Breurec S, Guessennd N, Timinouni M, Le TA, Cao V, Ngandjio A, Randrianirina F, Thiberge JM, Kinana A, Dufougeray A, Perrier-Gros-Claude JD, Boisier P, Garin B, Brisse S (2012) Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect. [Epub ahead of print]
Gangoué-Piéboji J (2007) Caractérisation des beta-lactamases et leur inhibition par les extraits de plantes médicinales. BICTEL/e—ULG Serveur institutionnel des thèses de doctorat
Gangoué-Piéboji J, Bedenic B, Koulla-Shiro S, Randegger C, Adiogo D, Ngassam P, Ndumbe P, Hächler H (2005) Extended-spectrum-beta-lactamase-producing Enterobacteriaceae in Yaounde, Cameroon. J Clin Microbiol 43(7):3273–3277
Jarlier V, Nicolas MH, Fournier G, Philippon A (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10(4):867–878
Clinical and Laboratory Standards Institute (CLSI) (2010) Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. CLSI document M100-S20. CLSI, Wayne, PA
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65(3):490–495
Pagani L, Dell’Amico E, Migliavacca R, D’Andrea MM, Giacobone E, Amicosante G, Romero E, Rossolini GM (2003) Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol 41(9):4264–4269
Bomasang ES, Mendoza MT (2003) Prevalence and risk factors associated with extended spectrum beta lactamase (ESBL) production among selected Enterobacteriaceae isolates at the Philippine General Hospital. Phil J Microbiol Infect Dis 32:151–158
Tonkic M, Goic-Barisic I, Punda-Polic V (2005) Prevalence and antimicrobial resistance of extended-spectrum beta-lactamases-producing Escherichia coli and Klebsiella pneumoniae strains isolated in a university hospital in Split, Croatia. Int Microbiol 8(2):119–124
Ramazanzadeh R, Mansouri M (2009) Spread of extended-spectrum beta-lactamase producing Escherichia coli clinical isolates in Sanandaj hospitals. J Biol Sci 9:362–366
Machado E, Coque TM, Cantón R, Novais A, Sousa JC, Baquero F, Peixe L; Portuguese Resistance Study Group (2007) High diversity of extended-spectrum beta-lactamases among clinical isolates of Enterobacteriaceae from Portugal. J Antimicrob Chemother 60(6):1370–1374
Severin JA, Mertaniasih NM, Kuntaman K, Lestari ES, Purwanta M, Lemmens-Den Toom N, Duerink DO, Hadi U, van Belkum A, Verbrugh HA, Goessens WH; Study Group ‘Antimicrobial Resistance in Indonesia: Prevalence and Prevention’ (AMRIN) (2010) Molecular characterization of extended-spectrum beta-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya, Indonesia. J Antimicrob Chemother 65(3):465–469
Ahoyo AT, Baba-Moussa L, Anago AE, Avogbe P, Missihoun TD, Loko F, Prévost G, Sanni A, Dramane K (2007) Incidence of infections dues to Escherichia coli strains producing extended spectrum betalactamase, in the Zou/Collines Hospital Centre (CHDZ/C) in Benin. Med Mal Infect 37(11):746–752
Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DS, Urassa WK, Fataki M, Msangi V, Tellevik MG, Maselle SY, Langeland N (2005) High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J Clin Microbiol 43(2):745–749
Seid J, Asrat D (2005) Occurrence of extended spectrum beta-lactamase enzymes in clinical isolates of Klebsiella species from Harar region, eastern Ethiopia. Acta Trop 95(2):143–148
Mirelis B, Navarro F, Miró E, Mesa RJ, Coll P, Prats G (2003) Community transmission of extended-spectrum beta-lactamase. Emerg Infect Dis 9(8):1024–1025
Smith DL, Dushoff J, Perencevich EN, Harris AD, Levin SA (2004) Persistent colonization and the spread of antibiotic resistance in nosocomial pathogens: resistance is a regional problem. Proc Natl Acad Sci USA 101(10):3709–3714
Lonchel CM, Meex C, Gangoué-Piéboji J, Boreux R, Okomo Assoumou MC, Melin P, De Mol P (2012) Proportion of extended-spectrum β-lactamase-producing Enterobacteriaceae in community setting in Ngaoundere, Cameroon. BMC Infect Dis 12(1):53
Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, Raz R (2004) Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis 23:163–167
Andriatahina T, Randrianirina F, Hariniana ER, Talarmin A, Raobijaona H, Buisson Y, Richard V (2010) High prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis 10:204
Calbo E, Romaní V, Xercavins M, Gómez L, Vidal CG, Quintana S, Vila J, Garau J (2006) Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum beta-lactamases. J Antimicrob Chemother 57(4):780–783
Rodríguez-Baño J, Navarro MD, Romero L, Martínez-Martínez L, Muniain MA, Perea EJ, Pérez-Cano R, Pascual A (2004) Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J Clin Microbiol 42:1089–1094
Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO (2001) Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis 32(8):1162–1171
Kuster SP, Hasse B, Huebner V, Bansal V, Zbinden R, Ruef C, Ledergerber B, Weber R (2010) Risks factors for infections with extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae at a tertiary care university hospital in Switzerland. Infection 38(1):33–40
Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, El-Defrawy I, Abou-Aitta A, Klena J, Nicolas-Chanoine MH (2011) CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb Drug Resist 17(1):67–73
Ben-Ami R, Schwaber MJ, Navon-Venezia S, Schwartz D, Giladi M, Chmelnitsky I, Leavitt A, Carmeli Y (2006) Influx of extended-spectrum beta-lactamase-producing Enterobacteriaceae into the hospital. Clin Infect Dis 42(7):925–934
Gangoué-Piéboji J, Koulla-Shiro S, Ngassam P, Adiogo D, Ndumbe P (2006) Antimicrobial activity against gram negative bacilli from Yaounde Central Hospital, Cameroon. Afr Health Sci 6(4):232–235
Peirano G, van Greune CH, Pitout JD (2011) Characteristics of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn Microbiol Infect Dis 69(4):449–453
Drieux L, Brossier F, Duquesnoy O, Aubry A, Robert J, Sougakoff W, Lecso-Bornet M, Jarlier V (2009) Increase in hospital-acquired bloodstream infections caused by extended spectrum beta-lactamase-producing Escherichia coli in a large French teaching hospital. Eur J Clin Microbiol Infect Dis 28(5):491–498
Silva J, Aguilar C, Becerra Z, López-Antuñano F, García R (1999) Extended-spectrum beta-lactamases in clinical isolates of enterobacteria in Mexico. Microb Drug Resist 5(3):189–193
Ho PL, Poon WW, Loke SL, Leung MS, Chow KH, Wong RC, Yip KS, Lai EL, Tsang KW; COMBAT study group (2007) Community emergence of CTX-M type extended-spectrum beta-lactamases among urinary Escherichia coli from women. J Antimicrob Chemother 60(1):140–144
Ndugulile F, Jureen R, Harthug S, Urassa W, Langeland N (2005) Extended spectrum beta-lactamases among Gram-negative bacteria of nosocomial origin from an intensive care unit of a tertiary health facility in Tanzania. BMC Infect Dis 5:86
Ehlers MM, Veldsman C, Makgotlho EP, Dove MG, Hoosen AA, Kock MM (2009) Detection of blaSHV, blaTEM and blaCTX-M antibiotic resistance genes in randomly selected bacterial pathogens from the Steve Biko Academic Hospital. FEMS Immunol Med Microbiol 56(3):191–196
Touati A, Brasme L, Benallaoua S, Madoux J, Gharout A, de Champs C (2008) Enterobacter cloacae and Klebsiella pneumoniae isolates producing CTX-M-15 recovered from hospital environmental surfaces from Algeria. J Hosp Infect 68(2):183–185
Touati A, Benallaoua S, Forte D, Madoux J, Brasme L, de Champs C (2006) First report of CTX-M-15 and CTX-M-3 beta-lactamases among clinical isolates of Enterobacteriaceae in Béjaia, Algeria. Int J Antimicrob Agents 27(5):397–402
Touati A, Benallaoua S, Djoudi F, Madoux J, Brasme L, De Champs C (2007) Characterization of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from hospital environments in Algeria. Microb Drug Resist 13(2):85–89
Olowe OA, Grobbel M, Büchter B, Lübke-Becker A, Fruth A, Wieler LH (2010) Detection of bla(CTX-M-15) extended-spectrum beta-lactamase genes in Escherichia coli from hospital patients in Nigeria. Int J Antimicrob Agents 35(2):206–207
Frank T, Arlet G, Gautier V, Talarmin A, Bercion R (2006) Extended-spectrum beta-lactamase-producing Enterobacteriaceae, Central African Republic. Emerg Infect Dis 12(5):863–865
Dahmen S, Bettaieb D, Mansour W, Boujaafar N, Bouallègue O, Arlet G (2010) Characterization and molecular epidemiology of extended-spectrum beta-lactamases in clinical isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb Drug Resist 16(2):163–170
Damjanova I, Tóth A, Pászti J, Bauernfeind A, Füzi M (2006) Nationwide spread of clonally related CTX-M-15-producing multidrug-resistant Klebsiella pneumoniae strains in Hungary. Eur J Clin Microbiol Infect Dis 25(4):275–278
Valenzuela de Silva EM, Mantilla Anaya JR, Reguero Reza MT, González Mejía EB, Pulido Manrique IY, Darío Llerena I, Velandia D (2006) Detection of CTX-M-1, CTX-M-15, and CTX-M-2 in clinical isolates of Enterobacteriaceae in Bogota, Colombia. J Clin Microbiol 44(5):1919–1920
Duval V, Maiga I, Maiga A, Guillard T, Brasme L, Forte D, Madoux J, Vernet-Garnier V, De Champs C (2009) High prevalence of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Bamako, Mali. Antimicrob Agents Chemother 53(11):4957–4958
Ramdani-Bouguessa N, Mendonça N, Leitão J, Ferreira E, Tazir M, Caniça M (2006) CTX-M-3 and CTX-M-15 extended-spectrum beta-lactamases in isolates of Escherichia coli from a hospital in Algiers, Algeria. J Clin Microbiol 44(12):4584–4586
Gangoué-Piéboji J, Miriagou V, Vourli S, Tzelepi E, Ngassam P, Tzouvelekis LS (2005) Emergence of CTX-M-15-producing enterobacteria in Cameroon and characterization of a blaCTX-M-15-carrying element. Antimicrob Agents Chemother 49(1):441–443
Kariuki S, Corkill JE, Revathi G, Musoke R, Hart CA (2001) Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob Agents Chemother 45(7):2141–2143
Mohamed Al-Agamy MH, El-Din Ashour MS, Wiegand I (2006) First description of CTX-M beta-lactamase-producing clinical Escherichia coli isolates from Egypt. Int J Antimicrob Agents 27(6):545–548
Acknowledgements
This study was performed at Yaounde Central Hospital, Ngaoundere Protestant Hospital and Ngaoundere Regional Hospital in Cameroon. We thank the authorities of these institutions for enabling us to conduct this study. We are grateful to the Chief Doctor, Simon Z. Aroga, who contributed to the study. We sincerely thank Moїse Woachie and all the laboratory technicians for their help in the collection of samples and in the interviews with the patients.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lonchel, C.M., Melin, P., Gangoué-Piéboji, J. et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in Cameroonian hospitals. Eur J Clin Microbiol Infect Dis 32, 79–87 (2013). https://doi.org/10.1007/s10096-012-1717-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-012-1717-4