Abstract
We report an outbreak of linezolid-resistant Staphylococcus haemolyticus strains (MIC 32 mg/L) in patients admitted to the Verona University Hospital Intensive Care Unit. The strains proved to be clonally related at pulsed field gel electrophoresis. All the strains showed the G2576T mutation responsible for linezolid-resistance and retained their resistance even after several passages on antibiotic-free medium. After a decade of linezolid use, multifocal emergence of linezolid resistance in coagulase-negative staphylococci has become an important matter of concern and mandates stricter control over the use of this antibiotic in order to preserve its clinical utility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococci with acquired multi-drug resistance associated with methicillin resistance are a major problem recently spreading from hospital- to community-acquired infections. The oxazolidinone antimicrobial compound linezolid is normally active against many drug-resistant Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).
Although even recent reports confirm that resistance to linezolid is minimal among coagulase-negative staphylococci (CoNS) [1, 2], a number of documented cases have occurred in Europe [3–5].
Linezolid binds to the domain V region of 23S rRNA and the effect of binding to 50S is inhibition of 70S formation. Even though the binding sites are similar, the action mechanism of oxazolidinones differs from those of all known protein synthesis inhibitors. Linezolid mainly works by binding the P site, thus inhibiting initiation complex formation and also translocation of peptydil-tRNA from the A site to the P site [6].
Decreased ribosomal affinity for the oxazolidinones due to mutations at 23S rRNA in the central loop of domain V is the main cause of bacterial resistance to these drugs [7–10]. Most mutations defined in various species and associated with linezolid resistance occur by a G to U substitution at position 2576 in the vicinity of the P site [7, 11]. Furthermore, mutation in the conserved region of L4 riboprotein encoded by the rplD gene conferred cross-resistance to oxazolidinones, macrolides and chloramphenicol in Streptococcus pneumoniae [12]. As a third possibility, a cfr-gene- encoded methyl-transferase, which alters adenosine at position 2503 in 23S rRNA, has also been reported in clinical strains [13, 14].
We report here on an outbreak of linezolid-resistant S. haemolyticus (MIC, 32 mg/L) isolated from patients admitted to the Verona University Hospital ICU.
Materials and methods
Patients and strains
The SHA 200 linezolid-resistant S. haemolyticus strain was isolated from a blood culture of a 65-year-old male admitted to the Verona University Hospital ICU on 1 August 2006, after extensive surgery for acute pancreatitis.
Empirical therapy with teicoplanin (600 mg) was started on admission followed by 200 mg/day after 48 h. On the sixth day of therapy, a strain of S. haemolyticus was isolated from the blood culture. The strain proved resistant to teicoplanin (MIC, 16 mg/L), which was immediately suspended and replaced with linezolid (MIC, 0.25 mg/L). After a further 8 days, a new blood culture showed that the same strain had become resistant to linezolid (MIC, 32 mg/L). The strain was investigated with a view to establishing its resistance mechanism.
We also included in the study two linezolid-susceptible strains of S. haemolyticus (SHA 203 and SHA 204) isolated from blood cultures of two different patients admitted to the ICU over the same period as the index case. All the strains were methicillin-resistant.
SHA 200 was also compared to ten additional linezolid-resistant strains of S. haemolyticus subsequently isolated in the same unit from non-duplicated patients over the following 6 months. Table 1 summarizes the clinical data for the isolates.
Antimicrobial susceptibility testing
The MICs of all antibiotics were determined by means of the E-test and confirmed by microdilution tests according to the latest EUCAST documents [15].
Stability of the resistant phenotype
To study the stability of the resistant phenotype in the absence of antimicrobial selective pressure, a single colony of the linezolid-resistant S. haemolyticus, SHA 200, was serially transferred 30 times on antibiotic-free medium (Müller-Hinton agar) incubated overnight at 37°C. The linezolid MICs for all organisms recovered after each of the 30 passages were determined by E-test.
Growth curves
The growth curves of linezolid-susceptible and linezolid-resistant strains were compared by measuring the optical density at 640 nm at one-hour intervals. The initial inocula were obtained by diluting the overnight cultures so as to achieve the same concentrations of the two strains.
Pulsed field gel electrophoresis (PFGE)
PFGE was carried out using standard procedures. The DNA preparation was digested with 30 U of SmaI for 3 h. The fragments were separated with a linear ramped pulse time of 6.8–63.8 over a period of 23 h at 14°C [16].
Detection of the G2576T mutation
Both the linezolid-susceptible and the linezolid-resistant strains were analyzed for the presence of the G2576T mutation (according to the Escherichia coli numbering system) in the V domain of the 23S rRNA gene. The primers used were 23Sfw and 23Srev [7] with the following cycling conditions: 30 cycles consisting of 94°C for 1 min, 50°C for 30 sec, and 72°C for 1 min.
The PCR fragments (596 pb) were purified and sequenced.
We also used a polymerase chain reaction (PCR) to check for the other known mechanisms of linezolid resistance, namely, the rplD (L4 ribosomal protein) and cfr (chloramphenicol florfenicol resistance) genes. All the oligonucleotide primers applied had been described previously [13, 17]. In the case of the rplD gene, the cycling parameters were the following: 30 cycles consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Amplification of the cfr gene was performed according to PCR parameters published previously [17]. The rplCfw primer was earlier published by Locke et al. [18], and the rplCrev primer (CGT AAT GAC GCA CGT TGT AAG: corresponding to 1861260–1861280 nucleotide bases of S. epidermidis RP62A, GenBank accession no. NC_002976) was designed on the MWG Operon homepage. For the amplification of the rplC gene, 33 cycles were applied with 94°C denaturation (1 min), 53°C annealing (1 min) and 72°C temperature (1 min).
Positive PCR products were purified (Qiagen) and sequenced (MWG Operon) on both strands. The DNA sequences obtained were aligned with the corresponding nucleotide sequences of the E. coli reference strain (GenBank accession no V00331) in NCBI Blast. The rplC and rplD gene sequence results were analysed on amino-acid level aligned with reference strain S. haemolyticus JCSC1435 DNA (L3 riboprotein ID: BAE04111.1 and L4 riboprotein ID: BAE04112.1).
Determination of number of alleles mutated
Five μl of PCR product were digested with 10 U of Nhe I restriction enzyme at 37°C for 1 h. The complete digestion of the PCR products indicated that all the alleles were mutated.
Results
Table 2 reports the MIC values of linezolid, vancomycin, teicoplanin and other recently released anti-staphylococcal compounds for SHA 200 (linezolid-resistant) and SHA 203 (linezolid-susceptible). The MIC values for all the additional linezolid-resistant isolates of S. haemolyticus were exactly the same as for SHA 200, and those for SHA 203 were exactly the same as for SHA 204.
After 30 passages on antibiotic-free medium, the resistance of strain SHA 200 was still unvaried at 32 mg/L.
Compared with the linezolid-susceptible strain, all the linezolid-resistant strains showed a significant difference in their in-vitro growth characteristics. Their growth rates were significantly slower, although at 24 h the OD640 nm was the same as for the susceptible strain.
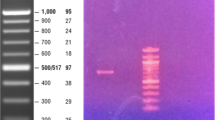
Figure 1 shows the PFGE results for the linezolid-susceptible strain, the linezolid-resistant strain SHA 200 and an additional 5 linezolid-resistant strains isolated between December 2006 and February 2007 in the Verona University Hospital ICU. All the resistant strains had the same pattern, which is clearly different from the susceptible strain SHA 203, thus proving the clonal relationship between the linezolid-resistant strains.
Pulsed field gel electrophoresis (PFGE) of clinical strains involved in the outbreak. Line 1 molecular weight; line 2 SHA200 index strain; line 3 SHA203 susceptible strain; line 4 SHA205; line 5 SHA206; line 6 SHA214; line 7 SHA219; line 8 SHA224 susceptible strain; line 9 SHA 226; line 10 SHA228; line 11 SHA233; line 12 SHA234; line 13 SHA239; line 14 SHA240
After genetic analysis, all the linezolid-resistant strains showed the G2576T mutation in domain V of 23S rRNA. The linezolid-susceptible strains did not present this mutation, which has been shown to be responsible for the emergence of linezolid resistance [7].
The cfr gene was not detected in any of the isolates with the previously published primer pairs [13, 17]. After amplification of rplC and rplD genes, the nucleic acid and amino acid sequences of amplicons showed the wild type of L3 and L4 riboprotein without any mutation as compared with the S. haemolyticus susceptible strain.
The complete digestion with the NheI of the PCR products compared with undigested PCR indicates that all the alleles were mutated.
Conclusions
After almost ten years of linezolid use, and although the overall prevalence of linezolid resistance is still low [1], multifocal emergence of linezolid resistance in CoNS in different geographical areas has become an important matter of concern and involves species other than S. epidermidis.
We reported an outbreak of linezolid-resistant S. haemolyticus in the Verona University Hospital, ICU with the index strain isolated from a blood culture of a 65-year-old male after empirical therapy with teicoplanin, and discovered the mechanism of resistance to be the classic mutation G2576T in domain V of 23S rRNA.
This finding confirms the emergence in Italy of CoNS clinical strains with resistance to linezolid and other second-line antibiotics, as well as with reduced susceptibility to glycopeptides. The outbreak described in this paper occurred in a different geographical area and is antecedent to those described in a recent report, which documented for the first time the isolation of linezolid-resistant CoNS in Italy and demonstrated different linezolid resistance mechanisms in multiple CoNS species, namely, S. epidermidis, S. hominis and S. simulans, but not S. haemolyticus [4]. Thus, it broadens the picture of linezolid-resistance in CoNS in terms of time of onset, geographical spread and species concerned. Whilst no link with antibiotic usage was observed in the report by Bongiorno et al. (2010), the present outbreak seems to confirm the close relationship between linezolid use and resistance.
S. haemolyticus, a frequent coloniser of human skin which is second in frequency only to S. epidermidis among clinical isolates of CoNS [19], has been regarded since the early studies as an important nosocomial pathogen with a tendency to develop multiple resistance [20]. Frequent insertion sequences in its chromosome probably account for frequent genomic rearrangements [21]. Indeed, it was the first Gram-positive pathogen to acquire glycopeptide resistance, and it has been suggested as being more active than other CoNS in generating clones with increased glycopeptide (especially teicoplanin) MICs [22].
The finding of linezolid-resistant strains in this species represents a disquieting addition to the landscape of antimicrobial resistance in Italy, and mandates stricter control over the use of linezolid to preserve its clinical utility.
References
Farrell DJ, Mendes RE, Ross J, Jones RN (2009) Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn Microbiol Infect Dis 65:392–403
Rossolini GM, Mantengoli E, Montagnani F, Pollini S (2010) Epidemiology and clinical relevance of microbial resistance determinants versus anti-Gram-positive agents. Curr Opin Microbiol 13:582–588
Kelly S, Collins J, Davin M, Gowing C, Murphy PG (2006) Linezolid resistance in coagulase-negative staphylococci. J Antimicrob Chemother 58:898–899
Bongiorno D, Campanile F, Mangelli G, Baldi MT, Provenzani R, Reali S, Lo Russo C, Santagati M, Stefani S (2010) DNA methylase modifications and other linezolid resistance mutations in coagulase-negative staphylococci in Italy. J Antimicrob Chemother 65:2236–2240
Rodriguez-Aranda A, Daskalaki M, Villar J, Sanz F, Otero JR, Chaves F (2009) Nosocomial spread of linezolid-resistant Staphylococcus haemolyticus infections in an intensive care unit. Diagn Microbiol Infect Dis 63:398–402
Bozdogan B, Appelbaum PC (2004) Oxazolidinones: activity, mode of action, and mechanism of resistance. Int J Antimicrob Agents 23:113–119
Tsiodras S, Gold SH, Saloulas G, Eliopoulos GM, Wennerste C, Venkataraman L, Moellering RC, Ferraro MJ (2001) Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207–208
Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB (2002) Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 46:3334–3336
Meka VG, Pillai SK, Sakoulas G, Wennersten C, Venkataraman L, De Girolami PC, Eliopoulos GM, Moellering RC, Gold HS (2004) Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J Infect Dis 190:311–317
Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA (2010) Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob Agents Chemother 54:742–748
Bonora MG, Solbiati M, Stepan E, Zorzi A, Luzzani A, Catania MR, Fontana R (2006) Emergence of linezolid resistance in the vancomycin-resistant Enterococcus faecium multi locus sequence typing C1 epidemic lineage. J Clin Microbiol 44:1153–1155
Wolter N, Smith AM, Farrell SJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP (2005) Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the Pneumococcus. Antimicrob Agents Chemother 49:3554–3557
Seok-Ming T, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS (2007) Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol 64:1506–1514
Mendes RE, Deshpande L, Rodriguez-Noriega E, Ross JE, Jones RN, Morfin-Otero R (2010) First report of staphylococcal clinical isolates in Mexico with linezolid resistance cause by cfr evidence of in vivo cfr mobilitation. J Clin Microbiol 48:3041–3043
The European Committee on Antimicrobial Susceptibility Testing (2011) Clinical breakpoints, version 1.1. Available at: http://www.eucast.org/clinical_breakpoints/. Accessed 19 July 2011
Shittu A, Lin J, Morrison D, Kolawole D (2006) Identification and molecular characterization of mannitol salt positive, coagulase-negative staphylococci from nasal samples of medical personnel and students. J Medic Microbiol 55:317–324
Kehrenberg C, Schwarz S (2006) Distribution of florfenicol resistance genes fexA and Cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163
Locke JB, Hilgers M, Shaw KJ (2009) Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother 53:5275–5278
Bannerman TL (2003) Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (eds) Manual of clinical microbiology, 8th edn. American Society for Microbiology, Washington, D.C.
Froggatt JW, Johnston JL, Galetto DW, Archer GL (1989) Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother 33:460–466
Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K (2005) Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187:7292–7308
Biavasco F, Vignaroli C, Varaldo PE (2000) Glycopeptide resistance in coagulase-negative staphylococci. Eur J Clin Microbiol Infect Dis 19:403–417
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mazzariol, A., Lo Cascio, G., Kocsis, E. et al. Outbreak of linezolid-resistant Staphylococcus haemolyticus in an Italian intensive care unit. Eur J Clin Microbiol Infect Dis 31, 523–527 (2012). https://doi.org/10.1007/s10096-011-1343-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-011-1343-6