Abstract
Antimicrobial resistance among community-acquired isolates of Escherichia coli is increasing globally, with international travel emerging as a risk for colonisation and infection. The aim was to determine the rate and duration of colonisation with resistant E. coli following international travel. One hundred and two adult hospital staff and contacts from Canberra, Australia, submitted perianal/rectal swabs before and following international travel. Swabs were cultured selectively to identify E. coli resistant to gentamicin, ciprofloxacin and/or third-generation cephalosporins. Those with resistant E. coli post-travel were tested monthly for persistent colonisation. Colonisation with antibiotic-resistant E. coli increased significantly from 7.8% (95% confidence interval [CI] 3.8–14.9) pre-travel to 49% (95% CI 39.5–58.6) post-travel. Those colonised were more likely to have taken antibiotics whilst travelling; however, travel remained a risk independent of antibiotic use. Colonisation with resistant E. coli occurred most frequently following travel to Asia. While over half of those carrying resistant E. coli post-travel had no detectable resistant strains two months after their return, at least 18% remained colonised at six months. Colonisation with antibiotic-resistant E. coli occurs commonly after international travel, and can be persistent. Medical practitioners should be aware of this risk, particularly when managing patients with suspected Gram-negative sepsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance among Escherichia coli is of increasing global concern [1–3]. This has been associated with the emergence and spread of extended-spectrum beta-lactamase (ESBL)-producing E. coli, which are also frequently associated with resistance to ciprofloxacin and aminoglycosides [4]. Surveillance data from intra-abdominal infections show rates of ESBL-producing E. coli of 42.2% in the Asia-Pacific, 21.6% in Latin America, 12.1% in Africa, 8% in Europe and 4.8% in North America [1, 4]. Within the Asia-Pacific, rates ranged from <5% in New Zealand to 55% in China and 79% in India [1]. Resistance rates of E. coli in Australia are low against third-generation cephalosporins (3GC) (1.5%), gentamicin (2.3%) and ciprofloxacin (3.4%) [5]. These antibiotics are considered to be “critically important” for human medicine [6]. Infections with resistant E. coli are traditionally associated with increasing age, hospitalisation, recent antibiotic use and chronic medical conditions [7, 8]. Some studies have also identified international travel as a risk factor for colonisation or infection with resistant E. coli [8–11], although only one study from 1983 [9] has prospectively studied faecal flora pre- and post-travel.
As resistant E. coli are readily acquired via ingestion [12, 13], we propose that, as people travel, their intestinal flora changes in response to the different strains of bacteria that they are exposed to via foods and water. Travel to regions with high rates of bacterial resistance, particularly those with sub-optimal sanitation and food and water supplies, should, therefore, be a risk factor for becoming colonised with resistant E. coli. On return to a region with low rates of antibiotic resistance, the clearance of resistant E. coli may occur as the “local” flora becomes re-established.
The aims of this study were to compare the rates of faecal colonisation with antibiotic-resistant E. coli in Australians pre- and post-travel, the duration of colonisation and the factors associated with colonisation with resistant E. coli.
Materials and methods
Setting, participants and design
The study was conducted in Canberra, Australia. Volunteers aged over 15 years of age who were due to travel internationally for at least one week were recruited via email among staff, and their contacts, of The Canberra Hospital. Participants were instructed to self-collect a rectal/perianal swab within two weeks of departure for international travel and within two weeks of their return to Australia. Questionnaires were used to collect demographic and travel information. “Antibiotic-resistant E. coli” was defined as E. coli resistant to one or more of the following antibiotics: gentamicin, 3GC or ciprofloxacin. Participants in whom resistant E. coli were identified post-travel submitted monthly rectal/perianal swabs until two consecutive swabs were negative, or for a maximum of six months, and were asked to report any infections during the six-month period. Travel occurred between January 2008 and April 2009, with follow-up completed in October 2009. The ACT Health Human Research Ethics Committee approved the study.
Microbiological methods
Swabs were inoculated into 2.5 ml of brain heart infusion broth containing a 30-μg vancomycin disk (Oxoid, UK), incubated overnight in room air at 35°C and then sub-cultured onto MacConkey agar, horse blood agar with gentamicin (HBA-gentamicin) (Oxoid, Australia) and chromID ESBL (bioMérieux, France). A 30-μg nalidixic acid (NA) disk (Oxoid, UK) was applied to the MacConkey agar. The plates were incubated in room air at 35°C for up to 48 h. Colonies typical of Enterobacteriaceae species on HBA-gentamicin and ESBL agars, or growing around the NA disk on the MacConkey agar, had identification and susceptibility testing performed using Vitek2 (bioMérieux, USA). E. coli non-susceptible to 3GC were tested phenotypically for AmpC β-lactamase production [14] and ESBL production [15]. ESBL and AmpC β-lactamase production was confirmed by polymerase chain reaction (PCR) tests as described for (TEM) [16], SHV [17], family-specific CTX-M [18] and plasmid-borne AmpC genes [19].
Statistical methods
Results were included in the data analysis only from participants who returned both pre- and post-travel swabs. McNemar’s or Fisher’s exact test, as appropriate, was used to assess for associations between categorical variables. A p-value of less than 0.05 was considered to be statistically significant and all tests were two-tailed. Analyses were performed using StataCorp 2009 statistical software (release 10.1, StataCorp, College Station, TX, USA).
Results
One hundred and six people were enrolled in the study and provided a pre-travel swab. Four participants (none of who had resistant E. coli isolated pre-travel) failed to return a post-travel swab and were excluded from further analysis. The median age was 45.4 years (range: 16.5–77.1 years), with 62% females. The median duration of travel was 21 days (range: 9–135 days). During the 12 months prior to departure, 32% of participants had travelled internationally, 15% had been hospitalised (including day units) and 43% had taken antibiotics. One-third of participants had immigrated to Australia, 85% arriving over 10 years ago and none within 12 months of travel.
E. coli accounted for over 92% of resistant Enterobacteriaceae isolated. Three gentamicin-resistant Klebsiella spp. and four Morganella spp. were also isolated. Pre-travel, 8 (7.8%) participants were colonised with resistant E. coli (Table 1). Each participant had only a single resistant strain of E. coli. Two participants were colonised with multi-resistant ESBL-producing E. coli (resistant to gentamicin, ciprofloxacin and 3GC). One had travelled to Eastern Europe/Middle East (CTX-M-1 group) two months previously and the other to India/Thailand and Western Europe (CTX-M-9 group) 12 months previously. Neither were colonised with the same strain post-travel. The first returned with an ESBL-negative gentamicin-resistant E. coli, whilst the second returned with a different multi-resistant ESBL-producing E. coli (CTX-M-1 group).
Immediately post-travel, 50 participants (49%) were colonised with at least one strain of resistant E. coli (Table 1). Twenty-four had one, 22 had two, one had three and three participants had four different resistant strains of E. coli isolated. Resistance to 3GC was due to the presence of ESBLs in 22 (21.6%) cases, predominantly CTX-M genotype (Table 2). Co-resistance to ampicillin (88%), amoxicillin-clavulanic acid (48%), cefazolin (46%) and co-trimoxazole (76%) was also common.
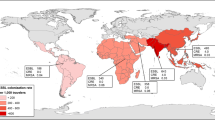
There was no difference in the median age (45.7 vs. 45.4 years), gender (60% vs. 63% female) or median duration of travel (21 vs. 21.5 days) between the groups returning with and without resistant E. coli. Participants with resistant E. coli were more likely to have developed gastroenteritis (52% vs. 23%, p < 0.005) or have taken antibiotics (38% vs. 17.3%, p < 0.05) whilst travelling. Antibiotics included doxycycline (9), norfloxacin (2), trimethoprim (1), azithromycin (1), tinidazole (1) and unknown (5) in those returning with resistant E. coli, and doxycycline (6), azithromycin (1) and amoxicillin (2) in those without resistant E. coli. The consumption of water that was not bottled or boiled was greater in those returning without resistant E. coli (79% vs. 46%, p < 0.01), and was still apparent when travellers to North America, Europe and Japan were excluded (21/30; 70% vs. 19/43; 44%, p = 0.03). Resistant E. coli was isolated from 50–79% of the participants who travelled to Asia (excluding Japan), South America and/or Middle East/Africa, but in less than 30% of those who travelled to Japan, North America and/or Europe (Table 3). The seven participants who returned with resistant E. coli after visiting North America and/or Europe had also travelled to Asia or the Middle East. Six were colonised with ESBL-producing E. coli (Table 2) and one with a ciprofloxacin-resistant strain after visiting the UK, Israel and UAE. Travel to the Indian sub-continent was associated with the highest rates of colonisation with ESBL-producing E. coli (8/14; 57%).
Of the 50 participants who returned with resistant E. coli, all but four completed the additional follow-up. Of those with incomplete follow-up, two participants with ciprofloxacin- and gentamicin-resistant E. coli, one with ciprofloxacin resistance and one with ciprofloxacin, gentamicin and 3GC resistance completed no, one, three and five months of follow-up, respectively. The majority of participants (26/48; 54%) cleared all resistant E. coli within two months; however, at least 18% of those returning with resistant E. coli remained persistently colonised at six months post-travel (Fig. 1). Clearance of E. coli resistant to 3GC was the most rapid, with only one (4%) participant colonised for more than three months. One woman developed a urinary tract infection with E. coli with identical antibiotic susceptibility to which she was colonised.
Discussion
Colonisation with E. coli resistant to gentamicin, ciprofloxacin and/or 3GC in Canberra residents pre-travel was low (7.8%) and similar to the low local rates of resistance seen among clinical isolates [5]; however, this rate rose markedly following international travel. Forty-nine percent carried at least one resistant isolate post-travel (Table 1), with 44% of these having multiple resistant strains. Colonisation with resistant strains of E. coli was temporary in the majority of these travellers, but did persist for more than six months in 18%, with another 8% having unknown status due to incomplete follow-up. Travel to the Indian sub-continent, China and the Middle East/Africa was associated with the highest rates of resistance in returned travellers. The consumption of antibiotics and the development of gastroenteritis during travel was more common in returned travellers colonised with resistant E. coli than travellers without resistant isolates. Paradoxically, travellers returning without resistant E. coli, however, were more likely to have consumed water that was not bottled or boiled. This may be explained by unmeasured confounders, such as the amount of water consumption, bottled versus boiled water and rural versus urban travel, but requires more detailed studies. It, however, may suggest that food is a more important source than water for acquiring these resistant bacteria. It is notable that one participant developed a clinical infection with a resistant E. coli with identical antibiotic susceptibilities to which they were colonised.
One other study has prospectively studied E. coli faecal flora in travellers [9], with our study being the only one to also document the duration of carriage with resistant strains. Rapid and multiple strain changes occurred in E. coli intestinal flora of Danes travelling to Egypt in the 1980s, with most new strains considered to be multi-resistant [9]. More recently, 24% of Swedish returning travellers with diarrhoea were colonised with ESBL-producing E. coli, with travel to the Middle East, India and Southeast Asia being the greatest risk [11]. Pre-travel colonisation rates were not studied; however, the results would be expected to be similar to the Australian rates. The association in our study with colonisation with resistant E. coli and gastroenteritis during travel is also consistent with the high rates of colonisation with ESBL-producing E. coli (41%) reported during an outbreak of salmonellosis [20], suggesting that the resistant E. coli are being ingested in contaminated food and/or water, along with enteric pathogens. Additional support to this is the recovery of ESBL-producing E. coli from vegetables, faeces of pigs, poultry and cattle, and sewage [21, 22]. Two retrospective studies have investigated the association between travel and infection with ESBL-producing E. coli. In New Zealand, 48.1% of people with a community-onset ESBL-producing E. coli urinary tract infection had recently travelled internationally, with most visiting India [10]. A Canadian study of community-onset ESBL-producing E. coli infections found that international travel was a significant risk factor for infection, with travel to India and the Middle East having the greatest risk [8]. Colonisation with resistant isolates in our study was more common in those visiting India and the Middle East/Africa, as well as China and Southeast Asia (Table 3), parallelling the prevalence of resistance seen in clinical isolates from these regions. Globally, community-onset ESBL-producing E. coli infections are most frequently associated with the CTX-M gene [2, 3, 23]. The CTX-M-1 group includes CTX-M-15, which originated in India, but is now found worldwide. Recent reports identifying the same CTX-M-15 clone (ST131) causing infections in different countries and continents supports the hypothesis that travel is an important factor in the dissemination of resistance [2, 24]. Of the E. coli resistant to 3GC in our study, the majority (18/26) were CTX-M ESBL-producers (Table 2), of which most belonged to the CTX-M-1 group, and occurred in travellers returning from India, Sri Lanka, Thailand and the Middle East. We also identified isolates belonging to the CTX-M-9 group. This is one of the most common groups occurring in China [23], and was seen in our study in travellers returning from China, Hong Kong and Vietnam.
There are some limitations of this study. The use of antibiotics was greater in travellers returning with resistant E. coli. The most common antibiotic used was doxycycline for malaria prophylaxis, with minimal use of fluoroquinolones and no use of 3GC or gentamicin. If all travellers who consumed antibiotics were excluded, 31 of 74 (42%) travellers returned with resistant E. coli, which remains a significant increase from the pre-travel rates (p < 0.0001). This indicates that, although a likely co-factor, antibiotic use alone was not responsible for the significant increase in colonisation with resistant E. coli post-travel. Travellers frequently visited multiple countries, making it difficult to determine the exact risk of acquiring resistant E. coli for each region. Although it is most likely that the resistant E. coli colonising the travellers who visited Europe and North America were acquired during their travel to Asia or the Middle East, it remains impossible to differentiate. Colonisation with resistant E. coli appeared temporary in most cases; however, it is possible that sub-populations of resistant E. coli may persist below the limit of detection, and predominate again if exposed to antibiotics. Although one participant developed an infection with a resistant E. coli, the study was not powered to address the correlation between colonisation and the subsequent risk of infection.
The results of our study suggest that resistant E. coli isolates are acquired from the environment during travel, presumably through food consumption, and that the acquisition of multiple different resistant strains is not uncommon. The likelihood of acquiring resistant organisms was greater in regions with known high rates of resistant E. coli, and may have been further increased by the more frequent use of antibiotics by travellers to these regions. Medical practitioners need to be aware of the association between travel and colonisation with multi-resistant bacteria, particularly in countries with low baseline levels of resistance, as this may impact clinical decisions concerning patient management, antibiotic choices and infection control practices. There are also important public health implications of rising antibiotic resistance, and this study has added further support to increasing evidence that travellers may be aiding in the spread of resistant E. coli. Although colonisation with resistant E. coli was temporary in most cases, at least 18% of those with resistant E. coli remained colonised at six months, raising concerns that these people will have ongoing personal risk of infection with resistant strains and also act as reservoirs for infection within the community.
The results from this study have identified several clinically significant questions requiring further research. What is the risk of infection following colonisation, and, hence, what antibiotics should be recommended for the empirical treatment of Gram-negative sepsis in a returned traveller? For how long post-travel should travellers be considered at risk and will antibiotic treatment result in the re-emergence of resistant isolates? How should elective procedures in returned travellers, such as transrectal prostate biopsies, be approached if there is a significant risk of post-procedure Gram-negative bacteraemia? Post-prostate biopsy bacteraemia with multi-resistant organisms is increasingly reported in the literature [25]. What is the role of food and water, as well as the use of antibiotics in food animals, in the promotion of resistant strains in humans? Ongoing epidemiological surveillance studies will be required to determine whether, in the era of unrestricted global movement, low-prevalence regions, such as Australia, Europe and North America, are able to prevent the importation and dissemination of resistant E. coli.
References
Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL (2009) Emergence of high levels of extended-spectrum-β-lactamase-producing Gram-negative bacilli in the Asia-Pacific region: data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program, 2007. Antimicrob Agents Chemother 53:3280–3284
Peirano G, Pitout JDD (2010) Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321
Cantón R, Coque TM (2006) The CTX-M β-lactamase pandemic. Curr Opin Microbiol 9:466–475
Hawser SP, Bouchillon SK, Hoban DJ, Badal RE (2009) In vitro susceptibilities of aerobic and facultative anaerobic Gram-negative bacilli from patients with intra-abdominal infections worldwide from 2005–2007: results from the SMART study. Int J Antimicrob Agents 34:585–588
Pearson J, Turnidge J, Franklin C, Bell J (2007) Prevalence of antimicrobial resistances in common pathogenic Enterobacteriaceae in Australia, 2004: report from the Australian Group on Antimicrobial Resistance. Commun Dis Intell 31:106–112
Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM (2009) World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis 49:132–141
Calbo E, Romaní V, Xercavins M, Gómez L, Vidal CG, Quintana S et al (2006) Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum β-lactamases. J Antimicrob Chemother 57:780–783
Laupland KB, Church DL, Vidakovich J, Mucenski M, Pitout JDD (2008) Community-onset extended-spectrum β-lactamase (ESBL) producing Escherichia coli: importance of international travel. J Infect 57:441–448
Stenderup J, Orskov I, Orskov F (1983) Changes in serotype and resistance pattern of the intestinal Escherichia coli flora during travel. Results from a trial of mecillinam as a prophylactic against travellers’ diarrhoea. Scand J Infect Dis 15:367–373
Freeman JT, McBride SJ, Heffernan H, Bathgate T, Pope C, Ellis-Pegler RB (2008) Community-onset genitourinary tract infection due to CTX-M-15-producing Escherichia coli among travelers to the Indian subcontinent in New Zealand. Clin Infect Dis 47:689–692
Tham J, Odenholt I, Walder M, Brolund A, Ahl J, Melander E (2010) Extended-spectrum beta-lactamase-producing Escherichia coli in patients with travellers’ diarrhoea. Scand J Infect Dis 42:275–280
Corpet DE (1988) Antibiotic resistance from food. N Engl J Med 318:1206–1207
Collignon P (2009) Resistant Escherichia coli—we are what we eat. Clin Infect Dis 15:202–204
Coudron PE (2005) Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J Clin Microbiol 43:4163–4167
Clinical and Laboratory Standards Institute (CLSI) (2008) Performance standards for antimicrobial susceptibility testing; 18th informational supplement. M100-S18. CLSI, Wayne, PA
Hanson ND, Thomson KS, Moland ES, Sanders CC, Berthold G, Penn RG (1999) Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J Antimicrob Chemother 44:377–380
Rasheed JK, Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J et al (1997) Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother 41:647–653
Pitout JD, Hossain A, Hanson ND (2004) Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol 42:5715–5721
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Prats G, Mirelis B, Miró E, Navarro F, Llovet T, Johnson JR et al (2003) Cephalosporin-resistant Escherichia coli among summer camp attendees with salmonellosis. Emerg Infect Dis 9:1273–1280
Mesa RJ, Blanc V, Blanch AR, Cortés P, González JJ, Lavilla S et al (2006) Extended-spectrum β-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage). J Antimicrob Chemother 58:211–215
Duan RS, Sit THC, Wong SSY, Wong RC, Chow KH, Mak GC et al (2006) Escherichia coli producing CTX-M β-lactamases in food animals in Hong Kong. Microb Drug Resist 12:145–148
Hawkey PM (2008) Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect 14(Suppl 1):159–165
Pitout JDD, Campbell L, Church DL, Gregson DB, Laupland KB (2009) Molecular characteristics of travel-related extended-spectrum-β-lactamase-producing Escherichia coli isolates from the Calgary Health Region. Antimicrob Agents Chemother 53:2539–2943
Hadway P, Barrett LK, Waghorn DJ, Hasan K, Bdesha A, Haldar N et al (2009) Urosepsis and bacteraemia caused by antibiotic-resistant organisms after transrectal ultrasonography-guided prostate biopsy. BJU Int 104:1556–1558
Acknowledgements
We would like to acknowledge The Canberra Hospital Private Practice Fund for providing financial support, Dr. Geethanie Fernando and the microbiology staff of ACT Pathology, Jan Bell of SA Pathology (Women’s and Children’s Hospital Adelaide) and Dr. Marian Currie of The Academic Unit of Medicine, Australian National University.
Role of funding source
The Canberra Hospital Private Practice Fund provided funding for the study. The study sponsor was not involved in the study design, collection of data, interpretation of data, writing of the report or decision to submit the paper for publication. The corresponding author had full access to all of the data and had final responsibility for the decision to submit the paper for publication.
Conflict of interest
Both KK and PC declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kennedy, K., Collignon, P. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur J Clin Microbiol Infect Dis 29, 1501–1506 (2010). https://doi.org/10.1007/s10096-010-1031-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-1031-y