Abstract
The purpose of this study was to use microbiological culture and bacterial 16S rRNA gene sequencing methods to detect transcriptionally active bacteria present on the surface of failed prosthetic hip joints removed during revision arthroplasty. Five failed prosthetic hip joints were sonicated to dislodge adherent bacteria and subjected to microbiological culture. Bacterial RNA was extracted from each sonicate, cDNA prepared by reverse transcription and the 16S rRNA gene amplified using universal primers. Polymerase chain reaction (PCR) products were cloned, assigned to distinct groups by restriction fragment length polymorphism (RFLP) analysis and one representative clone from each group was sequenced. Bacteria were identified by comparison of the obtained 16S rRNA gene sequences with those deposited in public access sequence databases. All five specimens were positive for the presence of bacteria by both culture and PCR. Culture methods identified species from eight genera. Molecular detection of transcriptionally active bacteria identified a wider range of species. A total of 42 phylotypes were identified, of which Lysobacter gummosus was the most abundant (31.6%). Thirty-four clones (14.5%) represented uncultivable phylotypes. No potentially novel species were identified. It is concluded that a diverse range of transcriptionally active bacterial species are present within biofilms on the surface of failed prosthetic hip joints.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The risk of infection is a major complication of total hip joint arthroplasty. The diagnosis and treatment of such infections presents a significant clinical and financial burden and also results in high rates of morbidity of patients requiring prosthetic hip joint replacements. This also presents a major challenge for orthopaedic surgeons to achieve advances in accurate diagnosis of infection, as well as developing safe and effective antimicrobial treatments [1, 2]. It has been estimated that over 50,000 total hip replacements are performed each year in the UK, with the rate of infection remaining unacceptably high [3]. Prosthetic joint infections of total hip arthroplasties (THA) reportedly occur with an incidence of 1.5% for the primary THA and 3.2% for the revision THA [4]. However, one study demonstrated that up to 15% of hip replacements failed due to bacterial infection [5]. These infections are characterised by biofilms, adherent communities of bacteria attached to the prosthetic hip joint components that are resistant to antibiotic challenge and host immunity [6].
The difficulty in isolating, by traditional culture methods, the bacterial species present in the biofilm on the surface of the prosthetic hip joint makes prediction of the true rate of infection difficult [7]. Conventional clinical methods for the detection and identification of bacteria implicated in these infections are based on the pre- and peri-operative samples and the subsequent culture of bacteria. This method is neither specific, as associated microbes are often skin flora that may be contaminants, nor sensitive, as multiple tissue samples are cultured [7]. These conventional methods also have a number of associated problems in the detection of joint infection. Reasons for this problem include strongly adherent bacteria in the biofilm and the presence of antibiotic-containing cement. The microbial yield can be improved by carrying out mild ultrasonication of the prosthesis to remove adherent microbes from the biofilm on the surface of the prosthetic hip joint [8–10]. There may also be a low yield of microbial growth due to the fact that the joint is infected with highly fastidious and non-cultivable or viable but non-cultivable bacteria that cannot be isolated by the standard techniques. We, along with others, have shown that this problem can be overcome by the use of culture-independent, molecular detection methods for the diagnosis of infecting bacteria on the surface of orthopaedic implants [9, 11–14]. Our previous study using such methods identified bacterial species from both clinically infected and clinically non-infected prosthetic hip joint specimens [9].
The aim of the current study was to identify transcriptionally active bacteria within the biofilms on the surface of failed prosthetic hip joints by using both conventional microbiological culture and culture-independent (bacterial 16S rRNA gene sequencing) methods. Our previous study analysed bacterial DNA for the identification of bacterial species present on the surface of failed prosthetic hip joints [9], an approach that also detects dead and moribund bacteria in addition to live bacteria. However, the identification of transcriptionally active bacterial species, i.e. those species which are alive, could provide information on the key species which may be of greatest importance in the infective process. In order to achieve this, bacterial mRNA was detected within the biofilm of the failed prosthetic hip joint, rather than DNA as in our first study. This approach has been used previously to identify transcriptionally active bacteria in the synovial tissue of patients with rheumatoid arthritis and other arthropathies [15, 16]. Conventional microbiological culture was also carried out in order to identify as many bacterial species as possible that are associated with prosthetic hip joint infections. The clinical relevance of this study is to improve our understanding of the likely source of infecting bacteria which, in turn, may lead to possible preventative treatment methods against such infections.
Materials and methods
Patient selection
Ethical approval was obtained from the Ethics Committee of the Southern General Hospital, Glasgow. Patients undergoing prosthetic hip joint revisions were recruited from those attending the Department of Orthopaedic Surgery at the Southern General Hospital, Glasgow. Each patient gave written informed consent to participate in the study.
Clinical samples and clinical data
Five prosthetic hip joint implants (four clinically infected, one aseptic loosening) were retrieved by surgeons from patients undergoing revision hip surgery at the Southern General Hospital, Glasgow, as previously described [9]. Demographic and clinical data for the five patients are presented in Table 1. All five cases were clinically and radiologically loose, with a varying risk of infection, as demonstrated by raised levels of the infection markers C-reactive protein and erythrocyte sedimentation rate. Following removal, the femoral and acetabular cup components of the failed prosthetic hip joint were placed into sterile plastic bags and immediately analysed. Pre- and peri-operative samples (hip joint aspirate, capsular fluid, acetabular membrane and femoral membrane) were also obtained from each patient for routine bacteriological analysis. During revision surgery, no prophylactic antibiotics were administered until the bacteriology samples had been obtained and the prosthesis had been removed. The antibiotic-loaded cement used at each primary revision was cefuroxime with gentamicin.
Processing of pre-operative and peri-operative samples
Pre-operative and peri-operative samples were processed and subjected to both aerobic and anaerobic culture as described previously [9].
Processing of prosthetic hip joint components
The femoral and acetabular components of the prosthetic hip joint were processed separately and adherent bacteria removed by mild sonication as described previously [9]. The bacterial pellets obtained following the sonication of each component were pooled and stored at −80°C until required for molecular analysis.
Microbiological culture
The bacterial pellets obtained from the sonicated components were pooled and subjected to culture as described previously [9]. Briefly, 10-fold serial dilutions to 10−6 were prepared and subjected to aerobic culture on Columbia agar containing 7.5% v/v defibrinated horse blood (5% CO2 at 37°C) and anaerobic culture on Fastidious Anaerobe agar (85% N2, 10% CO2 and 5% H2 at 37°C). Culture was also carried out on Skim milk agar, nutrient agar and CY-agar plates (5% CO2 at 30°C). Isolated bacteria were identified by 16S rRNA gene sequencing as described below.
RNA extraction
RNA was extracted from bacterial cells present in the pooled sonicate from the acetabular cup and femoral component of the failed prosthetic hip joints. The RNA was isolated using TRIzol® Reagent (Invitrogen, Paisley, UK) in accordance with the manufacturer’s instructions. The yield, concentration and purity of RNA were determined spectrophotometrically. The integrity of the representative RNA samples was determined by the resolution of rRNA by denaturing gel electrophoresis.
DNase treatment
The RNA prepared from the bacterial cells present in the sonicate was treated with DNase in order to eliminate any residual DNA following the RNA extraction process. To the total volume of RNA prepared from each sonicated sample, the following components were added: 1 × First Strand Buffer (Invitrogen), 14 U of DNase I (Invitrogen) and 2 U of RNaseOut™ Ribonuclease Inhibitor (Invitrogen). Each reaction was incubated at 37°C for 10 min, ethylenediaminetetraacetic acid (EDTA) added to a final concentration of 2.5 mM and incubation continued at 65°C for 15 min. The DNase-treated RNA samples were stored at −20°C until required.
cDNA synthesis
cDNA was prepared from the DNase-treated RNA extracted from the bacterial species present in the sonicate of the failed prosthetic hip joints. For each cDNA preparation, 2 μg of isolated RNA was required. To the appropriate volume of RNA, 150 ng of random hexamer primers (Invitrogen) and 4 mM dNTPs (New England Biolabs, Hitchin, UK) were added and the final mixture made up to 10 µl with water. The mix was incubated at 65°C for 10 min, followed by storage on ice for 5 min. To each cDNA preparation, the following components were added: 1 × First Strand Buffer (Invitrogen), 10 mM DTT (Invitrogen) and 40 U RNaseOut™ Ribonuclease Inhibitor (Invitrogen). The mix was then stored at room temperature for 2 min, followed by the addition of 200 U of SuperScript™ II Reverse Transcriptase (Invitrogen). The reactions were incubated at room temperature for 10 min, 42°C for 50 min and 15 min at 70°C. The reactions were then stored on ice, 20 U of RNase H (Invitrogen) added and incubated at 37°C for 20 min. For polymerase chain reaction (PCR), either undiluted cDNA or cDNA diluted 1:10 or 1:50 in H2O was added to each reaction. The prepared cDNA was stored at −20°C.
PCR
Universal primers were used to amplify bacterial 16S rRNA genes, with generated cDNA acting as a template. The primer sequences were 5′-AGA GTT TGA TCM TGG CTC AG-3′ (27f; Escherichia coli nucleotides 8–27) and 5′-GGG CGG WGT GTA CAA GGC-3′ (1387r; Escherichia coli nucleotides 1387–1404) (MWG Biotech, Milton Keynes, UK), where M = C or A and W = A or T, and give an expected amplification product of approximately 1,400 base pairs [17]. All PCR reactions were carried out in a total volume of 50 μl, comprising 5 μl of bacterial cDNA (neat, 1:10 or 1:50 dilutions) and 45 μl of reaction mixture containing 1 × PCR buffer (10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8), 1.0 U of Taq DNA polymerase (New England Biolabs), 0.2 mM dNTPs (New England Biolabs) and each primer at a concentration of 0.2 μM. PCR was carried out in an OmniGene thermal cycler (Hybaid, Teddington, UK). The PCR cycling conditions were as follows: (i) denaturation at 94°C for 5 min; (ii) 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min and extension at 72°C for 1.5 min; (iii) extension at 72°C for 10 min.
PCR quality control
When carrying out PCR, stringent procedures were employed to prevent contamination, as previously described [9, 18]. In order to rule out the possibility of a PCR product being generated from contaminating DNA within the bacterial cDNA samples, two further controls were also included: one contained extracted RNA and the other contained DNase-treated RNA as templates for PCR.
Cloning of 16S rRNA PCR products
PCR products were cloned into pGEM-T Easy cloning vector using the pGEM-T Easy Vector System I Kit (Promega, Southampton, UK) in accordance with the manufacturer’s instructions.
PCR amplification of 16S rRNA gene inserts
Following cloning of the 16S rRNA gene products amplified by PCR for each sample, 50 clones from each generated library were randomly selected. The 16S rRNA gene insert from each clone was amplified by PCR with the primer pair 5′-GCT ATT ACG CCA GCT GGC GAA AGG GGG ATG TG-3′ (M13FAP) and 5′-CCC CAG GCT TTA CAC TTT ATG CTT CCG GCA CG-3′ (M13RAP). The M13FAP binding site is located 32 base pairs upstream of the M13 Forward primer binding site and the M13RAP binding site is located 39 base pairs downstream of the M13 Reverse primer binding site in the pGEM-T Easy vector.
Restriction enzyme analysis
Selected clones from the libraries generated from the five prosthetic hip samples (234 clones in total) were subjected to restriction fragment length polymorphism (RFLP) analysis with the restriction enzymes RsaI and MnlI, as described previously [9]. For each library, clones were assigned distinct RFLP groups on the basis of the restriction profiles obtained.
DNA sequencing
The 16S rRNA gene of a single, representative clone from each RFLP group identified by restriction enzyme analysis was sequenced using the CycleReader™ Auto DNA Sequencing Kit (Fermentas Life Sciences) and IRD800-labelled M13 universal (-21) (5′-TGT AAA ACG ACG GCC ACT-3′) or 16S rRNA 357f (5′-CTC CTA CGG GAG GCA GCA G-3′) primer on a Primus96 DNA thermal cycler (MWG Biotech), as described previously [9].
Analysis of 16S rRNA gene sequences
Sequence data were compiled with LI-COR Base ImagIR 4.0 software, converted to FASTA format and compared with 16S rRNA gene sequences from the GenBank and EMBL sequence databases using the advanced gapped BLAST program, version 2.1 [19]. Clone sequences possessing at least 98% identity with a sequence in the databases were identified as being that species.
Results
Culture-dependent methods
Diagnostic bacteriology results for the pre-operative and peri-operative samples (hip joint aspirate, capsular fluid, acetabular membrane, femoral membrane) for each of the five cases studied is shown in Table 1. Bacteria were isolated in at least one of these samples in all five patients. All bacteria isolated were from the Staphylococcus genus.
Bacteria were isolated from the sonicate of each of the four clinically infected and one non-infected prosthetic hip joint samples by conventional microbiological culture. From the five prosthetic hip joints analysed, a total of 19 bacterial isolates were obtained and identified by 16S rRNA gene sequencing. Table 2 shows the identity of the isolates obtained, grouped according to genera. Eight different genera were identified, with species belonging to the Virgibacillus genus being the most prevalent and accounting for 36.8% of the isolates analysed. Other predominant genera included Staphylococcus (15.9%), Bacillus (10.5%), Leifsonia (10.5%) and Sphingomonas (10.5%).
The bacteria isolated were also identified to the species level using 16S rRNA sequencing (Table 3). Ten different species were identified and the most prevalent species were Virgibacillus halophilus (36.8%), Bacillus permians (10.5%), Leifsonia sp. PTX1 (10.5%) and Staphylococcus aureus (10.5%).
Culture-independent methods
A total of 234 clones from the five failed prosthetic hip joint samples were subjected to RFLP analysis. Since many RFLP groups contained multiple clones with the same restriction profiles, a single representative clone from each group was sequenced in order to avoid sequencing redundancy. A DNA sequence of at least 500 nucleotides was obtained for each clone. A total of 98 clones were sequenced.
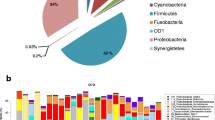
Twenty-three different bacterial genera/groups were identified in the five samples (Table 4). Lysobacter was the most predominant genus, accounting for 31.6% of the clones analysed. Other bacterial genera/groups identified included Staphylococcus (13.8%), uncultured bacterial clones (13.2%), Undibacterium (7.7%), Methylobacterium (6.8%) and Virgibacillus (5.5%). The bacteria identified were also categorised to the species level (Table 5). A total of 42 phylotypes were identified. Lysobacter gummosus was the most prevalent species, accounting for 31.6% of the clones analysed, followed by Staphylococcus aureus (12.4%), Undibacterium pigrum (7.7%), Methylobacterium sp. MP3 (6.8%) and Virgibacillus halophilus (5.5%).
Thirty-four (14.5%) analysed clones represented 17 different uncultured phylotypes (Table 6). The most prevalent phylotype was an uncultured environmental bacterium (GenEMBL accession number AB237728) previously isolated from fault-bordered aquifers [20], representing seven (3.0%) of the clones analysed. No potentially novel species (with sequence identities less than 98%) were identified.
Discussion
The main aim of this study was to identify transcriptionally active bacteria associated with failed prosthetic hip joints. Five prosthetic hip joint samples (four infected, one aseptic loosening) were sonicated to remove the bacterial cells within the adherent biofilm. The bacteria present in the sonicate were identified using both conventional microbiological culture and bacterial 16S rRNA gene sequencing using cDNA produced by reverse transcription of mRNA as a template. All five prosthetic hip joint samples were positive for the presence of bacteria by both detection methods. In our previous study [9], we used 16S rRNA gene sequencing to detect bacterial DNA in ten failed prosthetic hip joints (five infected, five aseptic loosening). A disadvantage of this molecular detection method is that the DNA of dead and moribund bacteria will be detected, in addition to those of live bacteria. This methodology was refined in the current study in that bacterial RNA was purified from the samples rather DNA and, following the preparation of cDNA by reverse transcription polymerase chain reaction (RT-PCR), was used as a template for 16S rRNA PCR. All five prosthetic hip joint samples gave positive results for RT-PCR following DNase treatment of the RNA, which provides evidence for the presence of transcriptionally active bacteria and eliminates the possibility of DNA contamination of the samples from spurious bacteria. The resultant 16S rRNA PCR products were cloned, sequenced and transcriptionally active bacteria present in each sample identified by BLAST analysis of obtained sequence data. We sought to minimise the sequencing of identical clones by screening cloned libraries using RFLP analysis and sequencing a single representative clone from each RFLP group. As described in our previous study [9], this is a common approach that has been used successfully in many studies to avoid sequencing redundancy and to estimate bacterial diversity within clinical specimens. Consequently, this allowed accurate relative quantification of the species present within each sample.
An understanding of which transcriptionally active bacteria are associated with failed prosthetic hip joints may ultimately aid in the reduction of infection rates and, subsequently, help to develop improved methods of prevention and treatment. There is still an ongoing debate over the source of bacteria which are capable of infecting prosthetic implants. The oral cavity can be a source of prosthetic joint infection [21–24], but the skin microbiota of the hospital staff or patients may also be a likely source of infecting bacteria. There is continuing uncertainty over the need for antibiotic prophylaxis when patients with joint prostheses undergo invasive dental treatment procedures [25, 26].
PCR amplification of the 16S rRNA gene has previously been shown to be used with great success for the identification of bacteria associated with prosthetic hip joint infections [12, 13, 27]. In contrast, some studies claim that PCR cannot be used to identify each pathogen in cases of mixed infection [28] and that it has poor positive predictive value for hip joint infection [29]. However, we have demonstrated both in our previous study [9] and in the current study that 16S rRNA gene amplification and sequencing is an extremely valuable approach for identifying bacterial species isolated by traditional microbiological culture methods and for defining the mixed bacterial population found on the surface of the prosthetic hip joints using a direct PCR and sequencing approach. It is important to note that we carried out PCR under stringent conditions and using appropriate controls to prevent false-positive results due to DNA contamination of RNA samples. Processing of the prosthetic hip joints and subsequent RNA extractions and cDNA synthesis were carried out in a separate laboratory from the PCR assays. All of the reagents for PCR were also stored separately from the positive cDNA samples with the reagents aliquoted before use to avoid contamination. Finally, a negative control was always included with each PCR assay to again rule out any possible contamination with bacterial DNA.
Previous research carried out by our group analysed the bacterial species from ten failed prosthetic hip joints using the same conventional culture and molecular diagnostic techniques [9]. The difference between these two studies is that, in the current study, we have analysed the bacterial mRNA rather than DNA to identify transcriptionally active bacteria thought to be involved in infection. This study has identified bacteria that are alive and functioning by analysing the species that are transcribing mRNA rather than also amplifying the DNA of dead or contaminating bacterial cells colonising the surface of the failed prosthetic hip samples. In our previous study, all ten cases were positive for the presence of bacteria by both culture and culture-independent methods. In that study, culture methods resulted in 46 bacterial isolates being recovered and were identified as species from the genera Leifsonia (54.3%), Staphylococcus (21.7%), Proteus (8.7%), Brevundimonas (6.5%), Salibacillus (4.3%), Methylobacterium (2.2%) and Zimmermannella (2.2%). Molecular detection methods identified a more diverse microflora. A total of 512 clones were analysed by RFLP analysis, of which 118 were sequenced. The predominant genus detected was Lysobacter, representing 312 (60.9%) of 512 clones analysed. A total of 28 phylotypes were identified: Lysobacter enzymogenes was the most abundant phylotype (31.4%), followed by Lysobacter sp. C3 (28.3%), gamma proteobacterium N4-7 (6.6%), Methylobacterium SM4 (4.7%) and Staphylococcus epidermidis (4.7%). Thirty-six clones (7.0%) represented uncultivable phylotypes. It is interesting to note in our current study that, in common with our previous study, a member of the Lysobacter genus (in this instance, Lysobacter gummosus) was the most prevalent transcriptionally active species. This reinforces our previous conclusion that Lysobacter species may play an important role in the infection.
From the cultured bacterial isolates sequenced, 19 different bacterial species were identified that are thought to be involved in prosthetic hip joint infections. Staphylococcus species have previously been described in these infections [8, 9, 27, 29], but the other bacteria identified are not commonly associated with prosthetic hip joint infections. The species identified by conventional culture methods from the surface of the failed prosthetic hip joints are, in most cases, environmental bacteria and, with the exception of Staphylococcus species, are not commonly associated with human disease. Virgibacillus species are spore-forming bacteria which have been previously isolated from soil [30] and Bacillus permians is an environmental bacterial species more commonly found in geological salt formations [31]. Leifsonia species are known to favour moist environments and, in association with other bacterial species, reportedly cause infections of the central venous catheter used as vascular access for haemodialysis [32]. Brevundimonas species are rarely isolated from clinical specimens and their role in human disease needs further investigation, although an association with two cases of bloodstream infections [33] and a case of septic arthritis in an immunocompetent child have been reported [34]. Brevundimonas and Dermacoccus species have also been identified by 16S rRNA gene sequencing as being part of a diverse range of bacterial species found in aortic aneurysms [35]. Conventional microbiological culture methods have also isolated Sphingomonas species in advanced noma lesions, infections which are open to the environment [36] and Peptoniphilus species have been isolated from human clinical specimens [37, 38].
Routine diagnostic bacteriology was also carried out on the pre-operative and peri-operative samples (hip joint aspirate, capsular fluid, acetabular membrane, femoral membrane) for each of the five cases studied. Staphylococcus species were found in all five cases, with Staphylococcus aureus specifically being identified in three of the cases (1, 2 and 4). Culture of the corresponding sonicates identified Staphylococcus aureus in cases 2 and 4, whereas Staphylococcus epidermidis was found in case 5. While these two sets of data are in general agreement in finding Staphylococcus species as the predominant cultivable organisms, the fact that these species were not found in sonicates from cases 1 and 3 can be attributed to the fact that different sample types were analysed and the spatial distribution of the organism may be different in these cases. However, the data clearly suggest that Staphylococcus species are the predominant cultivable organisms isolated from prosthetic hip joint sonicates and pre-operative/peri-operative samples. This finding is corroborated by the results of the culture-independent identification methods, which identified Staphylococcus aureus as the second most predominant species (12.4% of the total flora) in the sonicates. Since Staphylococcus aureus is considered to be a skin contaminant, one cannot rule out the possibility of the infection being introduced at the time of surgery.
In the current study, 42 bacterial phylotypes were identified by culture-independent molecular methods. The most prevalent transcriptionally active bacterial species identified on the surface of all failed prosthetic hip joints (both clinically infected and non-infected) was Lysobacter gummous. This supports the findings of our previous study [9], in which members of the Lysobacter genus were the most abundance species identified by culture-independent methods in the biofilm found on the surface of failed prosthetic hip joints. This finding, therefore, reinforces our previous conclusion that Lysobacter species may play an important role in the infection. However, despite the use of selective culture medium, Lysobacter species, which have not previously been reported to be involved in prosthetic hip infections, were not identified by microbiological culture methods in either of our studies. The role that Lysobacter-type species play in prosthetic hip joint infections is unknown and further research is required to study the virulence factors involved in infection and the effects on the human immune system. However, Lysobacter-type species have been shown to be important pathogens in hospital-acquired infections [39]. In fact, it has recently been demonstrated that various Lysobacter-type species have the ability to readily form biofilms on various substrates. These species include Stenotrophomonas maltophilia, Xylella fastidiosa and Xanthomonas axonopodis [40–42]. Therefore, it is unsurprising that these species were identified on the prostheses of the patients in our study.
Staphylococcus species were identified within the biofilm of prosthetic hip joints by both conventional culture and molecular techniques and have previously been reported to be involved in such infections [8, 9, 27, 29]. Other bacterial genera identified by both culture-dependent and culture-independent methods were Virgibacillus [30], Brevundimonas [33–35], Sphingomonas [36] and Peptoniphilus [37, 38]. Members of the Virgibacillus genus are environmental bacteria that have never been associated with human infections, whereas all the other genera identified by both methods have been isolated from human clinical specimens. Species belonging to the genera Dermacoccus and Bacillus were only identified by culture-dependent methods.
Several additional bacterial species were identified by culture-independent molecular diagnostic methods which were not isolated by culture methods. Undibacterium pigrum, which belongs to the family Oxalobacteraceae of the Betaproteobacteria, and Methylobacterium sp. have been isolated from drinking water [43] and shown to form biofilms with species from other distinct bacterial genera, including Staphylococcus [44]. Shigella flexneri is a human intestinal pathogen which is known to invade the epithelium of the colon and it has been reported that this type of enteric bacterium may play a role in the exacerbation of disease in patients with enthesitis-related arthritis [45]. Caulobacter species show similar taxonomy to Brevundimonas species [46], so may belong to the same genera and, as already discussed, Brevundimonas as well as other bacterial species including Dermacoccus and Bradyrhizobium have been identified within aortic aneurysms by 16S rRNA gene sequencing [35]. Escherichia coli is one of the many species living in the intestine of humans and can cause intestinal and extra-intestinal infections, including urinary tract infections, meningitis, peritonitis, mastitis, septicaemia and gram-negative pneumonia [47]. Escherichia coli has never been reported to be associated with hip joint infections, but its identification in our current study is not surprising due to its close association with other human infections. Acinetobacter infections have become common in hospitalised patients, especially in the intensive care unit [48], and it has been shown to be an emerging pathogen in patients with cystic fibrosis [49]. Rhodococcus species have been shown to cause infection in HIV-infected patients [50, 51].
Many of the uncultivable bacterial species identified in this present study are environmental bacteria [52–56] (Table 6). However, one such species has been identified in airway infections of intubated patients (GenEMBL accession number EF511932 [57]). The inability to culture these species by standard microbiological techniques in this study may have been due to the fastidious growth requirements of these organisms or due to these species existing within surface-associated biofilms in a viable but non-cultivable form. Many clinical and environmental bacteria are capable of this characteristic, but their pathogenicity remains largely unknown [58]. None of the patients in our current study received antibiotic prophylaxis before surgery, so this would not have hindered our ability to culture and isolate such bacteria.
The finding that Lysobacter gummosus was the predominant species (31.6% of clones analysed) identified in the prosthetic hip joint sonicates using culture-independent methods is particularly intriguing. Although regarded, in common with other members of the Lysobacter genus, as an environmental species, one cannot simply attribute this to contamination at the time of surgery or in the laboratory. The current study was carried out concomitantly alongside other distinct studies in our laboratory using identical methods and resources, but this species was never identified in any of our other studies. Therefore, consideration has to be given to the Lysobacter species as potential emerging pathogens. This situation potentially mirrors that of Stenotrophomonas maltophilia, a species formerly classified as a member of the Lysobacter genus [59] and first reported as an environmental species. However, Stenotrophomonas maltophilia is now known to be an important nosocomial pathogen that is associated with significant morbidity and mortality, especially in the immunocompromised and in cancer patients, and which can cause a variety of serious systemic infections, particularly bacteraemia and pneumonia [60, 61].
With the exception of our previous study [9], no other studies have associated Lysobacter species with the failure of prosthetic hip joints. However, using a direct PCR approach with species-specific primers, we have shown that Lysobacter enzymogenes and, indeed, Stenotrophomonas maltophilia are found at high levels (72.5 and 40.0%, respectively) on the surface of the human tongue (unpublished results). This suggests that the oral cavity may act as the source of infection for these two species and may help to explain the high incidence of these species on the surface of failed prosthetic hip joints in our previous study [9]. Given the findings of our current study, it would be prudent to investigate the prevalence of Lysobacter gummosus in the oral cavity in a similar manner.
In our current study, culture-independent methods identified several bacterial species thought to be involved in prosthetic hip joint infections. In our previous study in which we detected bacteria by the analysis of bacterial DNA rather than RNA, 28 distinct bacterial phylotypes were identified in a total of ten cases. In our current study analysing RNA of transcriptionally active bacteria, a total of 42 bacterial phylotypes were identified in a total of five cases. However, it should be noted that different patient samples were used in each study. Comparison of the results from both studies highlights an overlap of the main genera of bacteria isolated by culture-independent methods from the biofilms of failed prosthetic hip joints. The bacterial genera common to both studies are Lysobacter, Staphylococcus, Methylobacterium and Bradyrhizobium. A number of unusual bacterial species were identified in both studies which have not previously been described as human pathogens and which have not been implicated in the infection of failed prosthetic hip joints, and the majority of these species are environmental bacteria. Further research is required to study the pathogenicity of the bacterial species identified from the failed prostheses.
The culture-dependent and culture-independent identification methods used in this study showed some discordance in the types of bacteria identified. This demonstrates the necessity for using culture-independent methods in parallel with culture-dependent methods in order to identify the maximum number of bacteria associated with each failed prosthetic hip joint. The species which were successfully identified by both methods were from the genera Staphylococcus, Virgibacillus, Brevundimonas, Sphingomonas, Leifsonia and Peptoniphilus, of which only Staphylococcus has been previously associated with hip joint infections. As previously discussed [9], primer bias can occur during PCR, which results in the unequal amplification of certain PCR products. This will result in the preferential amplification of some bacterial DNA, leading to the under-representation of other species.
RT-PCR using universal primers to the bacterial 16S rRNA gene identified a wide range of bacterial species from the biofilm of failed prosthetic hip joints. No significant differences were observed in the bacterial species identified in the clinical situations of infection or aseptic loosening. Lysobacter gummosus was shown to be the most predominant transcriptionally active species. This study highlights the need for further research in this area, as a number of bacteria identified have not previously been reported to be involved in such infections. However, most of the bacterial species identified have been previously isolated from human clinical specimens. As a large number of species are now known to be involved in prosthetic joint infections, advances can be made in the study of bacterial biofilms and the pathogenic effect that these bacteria have on the human immune system. The key bacterial species involved in such infections can be further delineated by analysing a much larger number of samples. Quantitative studies using real-time PCR would aid in the determination of the actual abundance of specific infecting bacterial species associated with prosthetic hip joint infections.
In conclusion, a diverse range of transcriptionally active bacterial species were found to be present within biofilms on the surface of failed prosthetic hip joints. Virgibacillus halophilus was the most common species isolated by culture-dependent methods. Culture-independent methods identified Lysobacter gummosus and Staphylococcus aureus as the predominant transcriptionally active species.
References
Patel R, Osmon DR, Hanssen AD (2005) The diagnosis of prosthetic joint infection: current techniques and emerging technologies. Clin Orthop Relat Res 437:55–58
Toms AD, Davidson D, Masri BA, Duncan CP (2006) The management of peri-prosthetic infection in total joint arthroplasty. J Bone Joint Surg Br 88:149–155
NHS Direct Health Encyclopaedia. Hip replacement. Available online at: http://www.nhsdirect.nhs.uk/articles/article.aspx?articleId=522
Lentino JR (2003) Prosthetic joint infections: bane of orthopedists, challenge for infectious disease specialists. Clin Infect Dis 36:1157–1161
Lachiewicz PF, Rogers GD, Thomason HC (1996) Aspiration of the hip joint before revision total hip arthroplasty. Clinical and laboratory factors influencing attainment of a positive culture. J Bone Joint Surg Am 78:749–754
Thomas JG, Ramage G, Lopez-Ribot JL (2004) Biofilms and implant infections. In: Ghannoum M, O’Toole G (eds) Microbial biofilms. American Society of Microbiology Press, Washington, pp 269–293
Atkins BL, Athanasou N, Deeks JJ, Crook DWM, Simpson H, Peto TEA, McLardy-Smith P, Berendt AR (1998) Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. The OSIRIS Collaborative Study Group. J Clin Microbiol 36:2932–2939
Tunney MM, Patrick S, Gorman SP, Nixon JR, Anderson N, Davis RI, Hanna D, Ramage G (1998) Improved detection of infection in hip replacements. A currently underestimated problem. J Bone Joint Surg Br 80:568–572
Dempsey KE, Riggio MP, Lennon A, Hannah VE, Ramage G, Allan D, Bagg J (2007) Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res Ther 9:R46
Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R (2007) Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med 357:654–663
Clarke MT, Roberts CP, Lee PTH, Gray J, Keene GS, Rushton N (2004) Polymerase chain reaction can detect bacterial DNA in aseptically loose total hip arthroplasties. Clin Orthop Relat Res 427:132–137
Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, Gorman SP, Davis RI, Anderson N (1999) Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol 37:3281–3290
Mariani BD, Tuan RS (1998) Advances in the diagnosis of infection in prosthetic joint implants. Mol Med Today 4:207–213
Levine MJ, Mariani BA, Tuan RS, Booth RE Jr (1995) Molecular genetic diagnosis of infected total joint arthroplasty. J Arthroplasty 10:93–94
Cox CJ, Kempsell KE, Hill Gaston JS (2003) Investigation of infectious agents associated with arthritis by reverse transcription PCR of bacterial rRNA. Arthritis Res Ther 5:R1–R8
Kempsell KE, Cox CJ, Hurle M, Wong A, Wilkie S, Zanders ED, Hill Gaston JS, Crowe JS (2000) Reverse transcriptase-PCR analysis of bacterial rRNA for detection and characterization of bacterial species in arthritis synovial tissue. Infect Immun 68:6012–6026
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, pp 115–175
Riggio MP, Lennon A, Wray D (2000) Detection of Helicobacter pylori DNA in recurrent aphthous stomatitis tissue by PCR. J Oral Pathol Med 29:507–513
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Shimizu A, Akiyama M, Ishijima Y, Hama K, Kunimaru T, Naganuma T (2006) Molecular characterization of microbial communities in fault-bordered aquifers in the Miocene formation of northernmost Japan. Geobiol 4:203–213
Bartzokas CA, Johnson R, Jane M, Martin MV, Pearce PK, Saw Y (1994) Relation between mouth and haematogenous infection in total joint replacements. Br Med J 309:506–508
Stoll T, Stucki G, Brühlmann P, Vogt M, Gschwend N, Michel BA (1996) Infection of a total knee joint prosthesis by Peptostreptococcus micros and Propionibacterium acnes in an elderly RA patient: implant salvage with longterm antibiotics and needle aspiration/irrigation. Clin Rheumatol 15:399–402
Waldman BJ, Mont MA, Hungerford DS (1997) Total knee arthroplasty infections associated with dental procedures. Clin Orthop Relat Res 343:164–172
LaPorte DM, Waldman BJ, Mont MA, Hungerford DS (1999) Infections associated with dental procedures in total hip arthroplasty. J Bone Joint Surg Br 81:56–59
Tronstad L, Sunde PT (2003) The evolving new understanding of endodontic infections. Endod Top 6:57–77
Lockhart PB, Loven B, Brennan MT, Fox PC (2007) The evidence base for the efficacy of antibiotic prophylaxis in dental practice. J Am Dent Assoc 138:458–474
Fenollar F, Roux V, Stein A, Drancourt M, Raoult D (2006) Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 44:1018–1028
Zimmerli W, Trampuz A, Ochsner PE (2004) Prosthetic-joint infections. N Engl J Med 351:1645–1654
Panousis K, Grigoris P, Butcher I, Rana B, Reilly JH, Hamblen DL (2005) Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop 76:341–346
An SY, Asahara M, Goto K, Kasai H, Yokota A (2007) Virgibacillus halophilus sp. nov., spore-forming bacteria isolated from soil in Japan. Int J Syst Evol Microbiol 57:1607–1611
Graur D, Pupko T (2001) The Permian bacterium that isn’t. Mol Biol Evol 18:1143–1146
D’Amico M, Mangano S, Spinelli M, Sala E, Vigano EF, Grilli R, Fraticelli M, Grillo C, Limido A (2005) Epidemic of infections caused by ‘aquatic’ bacteria in patients undergoing hemodialysis via central venous catheters. G Ital Nefrol 22:508–513
Chi C-Y, Fung C-P, Wong W-W, Liu C-Y (2004) Brevundimonas bacteremia: two case reports and literature review. Scand J Infect Dis 36:59–61
Sofer Y, Zmira S, Amir J (2007) Brevundimonas vesicularis septic arthritis in an immunocompetent child. Eur J Pediatr 166:77–78
Marques da Silva R, Caugant DA, Eribe ERK, Aas JA, Lingaas PS, Geiran O, Tronstad L, Olsen I (2006) Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J Vasc Surg 44:1055–1060
Paster BJ, Falkler WA Jr, Enwonwu CO, Idigbe EO, Savage KO, Levanos VA, Tamer MA, Ericson RL, Lau CN, Dewhirst FE (2002) Prevalent bacterial species and novel phylotypes in advanced noma lesions. J Clin Microbiol 40:2187–2191
Wildeboer-Veloo ACM, Harmsen HJM, Welling GW, Degener JE (2007) Development of 16S rRNA-based probes for the identification of Gram-positive anaerobic cocci isolated from human clinical specimens. Clin Microbiol Infect 13:985–992
Song Y, Liu C, Finegold SM (2007) Peptoniphilus gorbachii sp. nov., Peptoniphilus olsenii sp. nov., and Anaerococcus murdochii sp. nov. isolated from clinical specimens of human origin. J Clin Microbiol 45:1746–1752
Looney WJ (2005) Role of Stenotrophomonas maltophilia in hospital-acquired infection. Br J Biomed Sci 62:145–154
Guilhabert MR, Kirkpatrick BC (2005) Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact 18:856–868
Jacques M-A, Josi K, Darrasse A, Samson R (2005) Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field-grown beans. Appl Environ Microbiol 71:2008–2015
Huang T-P, Somers EB, Wong ACL (2006) Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J Bacteriol 188:3116–3120
Kämpfer P, Rosselló-Mora R, Hermansson M, Persson F, Huber B, Falsen E, Busse HJ (2007) Undibacterium pigrum gen. nov., sp. nov., isolated from drinking water. Int J Syst Evol Microbiol 57:1510–1515
Simões LC, Simões M, Vieira MJ (2007) Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl Environ Microbiol 73:6192–6200
Saxena N, Misra R, Aggarwal A (2006) Is the enthesitis-related arthritis subtype of juvenile idiopathic arthritis a form of chronic reactive arthritis? Rheumatology 45:1129–1132
Abraham WR, Strömpl C, Meyer H, Lindholst S, Moore ER, Christ R, Vancanneyt M, Tindall BJ, Bennasar A, Smit J, Tesar M (1999) Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int J Syst Bacteriol 49:1053–1073
Smith JL, Fratamico PM, Gunther NW (2007) Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 4:134–163
Falagas ME, Karveli EA, Kelesidis I, Kelesidis T (2007) Community-acquired Acinetobacter infections. Eur J Microbiol Infect Dis 26:857–868
Davies JC, Rubin BK (2007) Emerging and unusual gram-negative infections in cystic fibrosis. Semin Respir Crit Car Med 28:312–321
Torres-Tortosa M, Arrizabalaga J, Villanueva JL, Gálvez J, Leyes M, Valencia ME, Flores J, Peña JM, Pérez-Cecilia E, Quereda C (2003) Prognosis and clinical evaluation of infection caused by Rhodococcus equi in HIV-infected patients: a multicenter study of 67 cases. Chest 123:1970–1976
Rozsypal H, Aster V, Stahiková M, Horová B (2007) Rhodococcus equi infection in subjects infected with human immunodeficiency virus (HIV). Cas Lek Cesk 146:163–167
North NN, Dollhopf SL, Petrie L, Istok JD, Balkwill DL, Kostka JE (2004) Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microbiol 70:4911–4920
Huber JA, Johnson HP, Butterfield DA, Baross JA (2006) Microbial life in ridge flank crustal fluids. Environ Microbiol 8:88–99
Kaiser O, Pühler A, Selbitschka W (2001) Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb Ecol 42:136–149
Rawls JF, Mahowald MA, Ley RE, Gordon JI (2006) Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433
Williams MM, Domingo JWS, Meckes MC, Kelty CA, Rochon HS (2004) Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J Appl Microbiol 96:954–964
Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Brown R, Hugenholtz P, DeSantis TZ, Andersen GL, Wiener-Kronish JP, Bristow J (2007) Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol 45:1954–1962
Oliver JD (2005) The viable but nonculturable state in bacteria. J Microbiol 43 Spec No:93–100
Giesler LJ, Yuen GY (1998) Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Prot 17:509–513
Senol E (2004) Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J Hosp Infect 57:1–7
Denton M, Kerr KG (1998) Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev 11:57–80
Acknowledgements
We thank Dr. Dominic Meek for the provision of the prosthetic hip joint samples and Dr. Grace Sweeney for conducting bacteriology on the pre-operative and peri-operative samples. This research was funded by the Arthritis Research Campaign (grant number 16418).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Riggio, M.P., Dempsey, K.E., Lennon, A. et al. Molecular detection of transcriptionally active bacteria from failed prosthetic hip joints removed during revision arthroplasty. Eur J Clin Microbiol Infect Dis 29, 823–834 (2010). https://doi.org/10.1007/s10096-010-0934-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-0934-y