Abstract
Eight patients with invasive bacteremic community-acquired methicillin-resistant Staphylococcus aureus infection in southeast Queensland, Australia, are reported. One patient died of septic shock. Haematogenous seeding to lungs, bone, and other sites was common. All isolates carried the virulence factor Panton-Valentine leukocidin and were either the southwest Pacific clone or the newly described Queensland clone. Clinicians should consider community-acquired methicillin-resistant Staphylococcus aureus infection in any patient presenting to hospital with severe staphylococcal sepsis or pneumonia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infections caused by methicillin-resistant Staphylococcus aureus (MRSA) have occurred throughout the world, primarily in healthcare settings [1]. Unfortunately, over the last 10 years, there have been increasing reports of community-acquired MRSA (CA-MRSA) infection [2–5]. The two common CA-MRSA strains in Australia are WA-MRSA, first described in Aboriginal communities of Western Australia [5], and the southwest Pacific strain, found in Pacific Islanders along the eastern seaboard [3]. More recently, CA-MRSA infections have emerged outside these defined risk groups, with a recent report of a new strain in Caucasians (Queensland strain) [6].
The clinical spectrum of CA-MRSA infections resembles those caused by methicillin-susceptible Staphylococcus aureus, with skin and soft tissue infections predominating. More recently, the invasive nature of CA-MRSA infection has been illustrated with increasing reports of necrotizing pneumonia [7, 8] and death [4]. The presumed hypervirulence of this organism has been linked to characteristic virulence factors, particularly Panton-Valentine leukocidin (pvl) which is encoded by the lukS-PV and lukF-PV genes. These genes code for a two-component bacterial toxin that leads to lysis of leukocytes and is thought to represent a stable genetic marker of CA-MRSA isolates [9].
Until now, invasive bacteremic CA-MRSA infection has been reported infrequently [2, 4, 6]. We have observed an increasing incidence of this presentation and, following the death of a young patient with septic shock, undertook this study to assess the demographic, clinical and microbiological features of life-threatening CA-MRSA infection in southeast Queensland, Australia.
Materials and methods
We performed a retrospective chart review of all patients who presented with bacteremic CA-MRSA infection to three large hospitals in southeast Queensland: Royal Brisbane and Women’s Hospital (January 2003–December 2003), Royal Children’s Hospital, Brisbane (January 2003–December 2003) and Ipswich Hospital (March 2000–December 2003). Cases were identified from MRSA blood culture isolates at each hospital’s microbiology department. All isolates that were obtained within 48 h of hospital admission [10] were followed up by consultation with the patient’s clinician, from whom the mode of acquisition was ascertained. Cases that were clearly hospital acquired were excluded. Patients were classified as having CA-MRSA infection with and without risk factors [11]. Risk factors included hospitalisation, outpatient visits, nursing home admission and antibiotic exposure within the previous 12 months. Other risk factors included chronic illness (diabetes mellitus, end-stage renal disease and malignancy), intravenous drug use and close contact with a person with furunculosis or risk factors for MRSA acquisition.
Antimicrobial susceptibility testing was performed on all isolates using the Vitek GPS-IX card (bioMérieux-Vitek, Hazelwood, MO, USA) and was confirmed using standard National Committee for Clinical Laboratory Standards disk methodology [12, 13]. Detection of mecA and lukF-PV genes was performed following extraction of genomic DNA as described by Huygens et al. [14]. The oligonucleotide primers for mecA detection were as described by Oliveira et al. [15]. The primers used for lukF-PV detection were designed according to the published sequences of the pvl genes (GenBank accession numbers X72700 and AB006796). The primer sequences for the lukF-PV gene were as follows: PVL_F1, 5′-TCACAAAATGCCAGTGTTATCCA-3′ and PVL_R1, 5′-TTTTGCAGCGTTTTGTTTTCG-3′. Polymerase chain reaction amplifications were performed as previously described [14], except for the lukF-PV gene cycling conditions, where an annealing temperature of 60°C and an extension time of only 30 s was used. Polymerase chain reaction products were resolved by electrophoresis and visualized with ethidium bromide. Pulsed-field gel electrophoresis of chromosomal DNA was performed according to standard methods [6].
The above research complies with the current laws of Australia.
Results
A total of 53 patients had an episode of MRSA bacteremia during the study period. Of these, 8 (15%) were identified as having CA-MRSA infection (Table 1). The median age was 18.5 years, and all patients were unrelated and Australian born. Four patients had no risk factors for acquiring MRSA. The remaining four patients had CA-MRSA with risk factors that included recent antibiotic exposure (case 2), monthly outpatient intravenous immunoglobulin for common variable immunodeficiency (case 3), diabetes mellitus (case 4) and intravenous drug use (case 6). No patients were hospitalised in the 12 months prior to admission.
All isolates were positive for the mecA and lukF-PV genes and were susceptible in vitro to all non-β-lactam antibiotics tested (erythromycin, clindamycin, gentamicin, ciprofloxacin, tetracycline, vancomycin, rifampicin and fusidic acid). Typing by pulsed-field gel electrophoresis showed two distinct clones of CA-MRSA (Fig. 1). The first was the southwest Pacific clone (cases 1, 2, 3 and 7), most commonly found in Pacific Islanders. One of the patients with this clone was of Pacific Island origin, two were Caucasian and one was Aboriginal. The second clone was the “R” pulsotype or Queensland clone (cases 4, 5, 6 and 8), identified in four patients (3 Aboriginal, 1 Caucasian). This is a new clone, recently reported in Queensland [6]. As previously published, the multilocus sequence types of the southwest Pacific and the Queensland clones are ST30 and ST93, respectively [9, 14]. Epidemiological links were observed in two patients (cases 7 and 8), both living in the same community.
Pulsed-field gel electrophoresis analysis of bacterial strains isolated from patients with life-threatening infection due to community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA). Lane 1 Gene Path control strain, Staphylococcus aureus 8325 (BioRad, Hercules, CA, USA); lane 2 size marker; lanes 3–5 and lane 9 cases 1–3 and 7, Southwest Pacific pulsotype of CA-MRSA; lanes 6–8 and lane 10 cases 4–6 and 8, R pulsotype (Queensland clone) of CA-MRSA
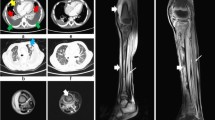
The source of bacteremia was clinically evident in seven patients, all being skin. One patient, case 5, had no identifiable source. Hematogenous seeding of peripheral sites was common (Table 1). This included: septic arthritis of the right knee requiring multiple surgical washouts (case 1); osteomyelitis involving a third metatarsal (case 2) and a left tibia (case 7), both requiring debridement; a deep pelvic collection (case 3); right-sided pyelonephritis with an upper pole collection (case 4); and deep soft tissue collections, including a left psoas and iliacus abscess (case 6) and a left sacral paraspinal abscess (case 8), both requiring surgical drainage. All patients had echocardiograms without evidence of endocarditis. Two patients had severe, necrotizing pneumonia requiring intubation and mechanical ventilation. One of these patients (case 5), a previously well 21-year-old Aboriginal male, presented in septic shock and died within 48 h of hospital admission [16]. He had a 1-week history of a progressive respiratory illness, including a productive cough, hemoptysis, dyspnoea and fevers. Initial examination revealed marked hypoxia (oxygen saturation of 79% breathing room air), a systolic blood pressure of 80 mm Hg, a temperature of 38.7°C and diffuse coarse crepitations. Chest radiograph revealed bilateral, multilobar consolidation with cavitation. Despite empirical antibiotic treatment (ceftriaxone, erythromycin, gentamicin and rifampicin) and maximal resuscitative efforts, the patient died. Sputum and blood cultures revealed Staphylococcus aureus, but susceptibility data were not available until after death. The second patient (case 6), a 34-year-old female with a history of IVDU, presented with fevers, left hip pain and a right forearm cutaneous abscess. Blood cultures were taken, and a computed tomography scan revealed a left psoas abscess. Within 48 h the patient developed tachypnoea, hypoxia and widespread inspiratory crepitations. This led to intubation and mechanical ventilation. A chest computed tomography scan revealed a cavitatory, necrotizing pneumonia. The patient made a slow recovery.
Five patients were treated with β-lactam antibiotics alone in the first 48 h. After the diagnosis of MRSA bacteremia was made, all patients apart from the patient who died were changed to intravenous vancomycin for a minimum of 4 weeks (2 patients with concurrent oral rifampicin). This was followed with oral clindamycin or combination therapy with rifampicin and fusidic acid for at least 2 weeks. Excluding the patient with the rapidly fatal outcome, all patients had prolonged hospital admissions with a median of 39 days (range, 19–82 days).
Discussion
CA-MRSA has emerged as a serious threat with important public health and clinical consequences. First, CA-MRSA is resistant to β-lactam antibiotics, the usual empiric therapy for suspected staphylococcal sepsis. As most infections with CA-MRSA involve skin and soft tissue and often respond to drainage alone, the impact of inappropriate empiric antibiotics has been low [6]. With the emergence of invasive life-threatening disease, antibiotic therapy has a more critical role. Our study illustrates the importance of recognizing previously described risk factors for CA-MRSA infection (intravenous drug use, Pacific Islander ethnicity) as well as new risk groups that have not been well described previously (patients of Aboriginal origin from southeast Queensland). Clinicians should be aware of the local prevalence of CA-MRSA so that demographic groups with increased risk receive appropriate empiric therapy on presentation with suspected staphylococcal septicemia or severe pneumonia.
Secondly, many strains of CA-MRSA have increased pathogenicity mediated by several virulence factors, particularly Panton-Valentine leukocidin, which was found in all of our isolates. Hematogenous seeding to joints, bone, lungs and deep soft tissue was common in our patients, with most requiring multiple surgical interventions and prolonged hospital admissions. Although necrotizing pneumonia in association with Panton-Valentine leukocidin has been reported for other strains of CA-MRSA [4, 7, 8], this is the first time it has been reported for the Queensland clone. Moreover, bacteremic CA-MRSA infection in patients of Aboriginal origin has not been well reported, possibly because most CA-MRSA infections in Australian Aboriginals are caused by WA-MRSA strains, which lack Panton-Valentine leukocidin and are hence less virulent than the Queensland clone [17].
This is the first observational series of bacteremic CA-MRSA infection in Queensland, Australia. CA-MRSA infections can cause significant morbidity and mortality in previously healthy young people, and rates of complicated bacteremia with hematogenous seeding of other sites is high. Clinicians should consider the possibility of CA-MRSA infection in any patient presenting to hospital with severe staphylococcal sepsis or pneumonia, particularly in geographic locations where CA-MRSA has been reported and in ethnic groups with increased risk.
References
Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M (2001) Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 32 (Suppl 2):114–132
Dufour P, Gillet Y, Bes M, Lina G, Vandenesch F, Floret D, Etienne J, Richet H (2002) Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin Infect Dis 35:819–824
Collignon P, Gosbell I, Vickery A, Nimmo G, Stylianopoulos T, Gottlieb T (1998) Community-acquired meticillin-resistant Staphylococcus aureus in Australia. Australian Group on Antimicrobial Resistance. Lancet 352:145–146
Centers for Disease Control and Prevention (1999) Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. JAMA 282:1123–1125
Riley TV, Rouse IL (1995) Methicillin-resistant Staphylococcus aureus in Western Australia, 1983–1992. J Hosp Infect 29:177–188
Munckhof WJ, Schooneveldt J, Coombs GW, Hoare J, Nimmo GR (2003) Emergence of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection in Queensland, Australia. Int J Infect Dis 7:259–264
Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J (1999) Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132
Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, Vandenesch F, Piemont Y, Brousse N, Floret D, Etienne J (2002) Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759
Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J (2003) Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 9:978–984
Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM (1988) CDC definitions for nosocomial infections, 1988. Am J Infect Control 16:128–140
Salgado CD, Farr BM, Calfee DP (2003) Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis 36:131–139
National Committee for Clinical Laboratory Standards (2000) Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A7. NCCLS, Villanova, PA
National Committee for Clinical Laboratory Standards (2001) Performance standards for antimicrobial disk susceptibility tests. Approved standard M100-S11. NCCLS, Villanova, PA
Huygens F, Stephens AJ, Nimmo G, Giffard PM (2004) mecA locus diversity in MRSA isolates in Brisbane, Australia, and the development of a novel diagnostic test for the Western Samoan Phage Pattern clone. J Clin Micro 42:1947–1955
Oliveira DC, Wu SW, de Lencastre H (2000) Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphisms of this region. Antimicrob Agents Chemother 44:1906–1910
Peleg AY, Munckhof WJ (2004) Fatal necrotising pneumonia due to community-acquired methicillin-resistant Staphylococcus aureus (MRSA). Med J Aust 181:228–229
Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O’Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitom C, Ito T, Hiramatsu K (2002) Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol 40:4289–4294
Acknowledgments
We thank the Ipswich Hospital Foundation and the Cooperative Research Centre for Diagnostics for funding this research. We are grateful to the microbiology department staff at Royal Brisbane and Women’s Hospital and Ipswich Hospital for collecting and storing the isolates. We also thank the physicians who cared for the patients during their inpatient admissions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peleg, A.Y., Munckhof, W.J., Kleinschmidt, S.L. et al. Life-threatening community-acquired methicillin-resistant Staphylococcus aureus infection in Australia. Eur J Clin Microbiol Infect Dis 24, 384–387 (2005). https://doi.org/10.1007/s10096-005-1329-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-005-1329-3