Abstract
Eukaryotic initiation factors 3 (EIF3) complex is essential for initiation of protein synthesis for both cells and virus. It consists of 13 subunits (EIF3A to M), among which EIF3B serves as a major scaffolding subunit. However, its functions in human glioblastoma have not been explored yet. Here, we showed that EIF3B was expressed in human glioblastoma (Grade I–IV) and human glioblastoma cell lines (U251, U87, A172 and U373). Loss of function analysis was performed on U87 cells using lentivirus-mediated siRNA against EIF3B. EIF3B-shRNA expressing lentivirus could effectively infect U87 glioma cells and downregulate EIF3B expression. Knockdown of EIF3B expression significantly inhibited proliferation of U87 cells. Further study showed that the proliferation inhibitory effect was associated with accumulation in G0/G1-phase cell number and an increased rate of apoptosis. In conclusion, EIF3B promotes the proliferation of U87 cells and may play an important role in human glioblastoma development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma is one the most malignant and common type of brain tumors with devastating outcome. Despite considerable advances in surgery, radiation, and chemotherapy, the prognosis for glioblastoma has not been substantially improved [1]. Because of the restricted anatomical location and absence of metastases outside the central nervous system (CNS) for glioblastoma, targeted gene therapy has shown a promising future in treating the disease [2]. Therefore, identifying target genes responsible for the tumorigenicity of glioblastoma will be useful for developing target therapy to treat glioblastomas.
Eukaryotic translation initiation factors (EIFs) were required to assemble 80S ribosomes onto mRNA with the initiator methionyl-tRNA (Met-tRNA) when protein synthesis happens [3]. The largest factor of these EIFs, EIF3, consists of 13 subunits (EIF3A to M) and with a total molecular mass of 700 kDa, plays an essential role in cellular and viral initiation of translation [4]. Among the subunits of EIF3, EIF3B is considered to be the major scaffolding subunit, interacting with EIF3A, G, I, and J [5, 6]. The human EIF3B is an 814-amino acid protein with an RNA recognition motif (RRM) located in its N terminus. The RRM domain could provide a specific platform for the interaction with EIF3J [7, 8]. Although the crystal structure of EIF3B has been analyzed, whether EIF3B plays a role in the tumorigenisis of glioblastomas has not been studied yet.
In the present study, we used the lentivirus expression system to knockdown EIF3B expression in U87 glioblastoma cells. Knockdown of EIF3B expression dramatically inhibited the proliferation ability of U87 cells, suggesting that EIF3B may function as an oncogene.
Experimental procedures
Materials
A total of ten freshly resected human glioblastoma specimens from Daping Hospital of the Third Military Medical University were analyzed. The study was approved by the ethics committee at the Third Military Medical University. Among the ten human glioblastoma specimens, two cases were Grade I, two were Grade II, three were Grade III, and three were Grade IV.
U87 glioblastoma cell line and HEK293T cell line were purchased from ATCC and maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 50 μg/ml streptomycin at 37°C in humidified CO2 incubator (5% CO2, 95% air). pGCSIL-GFP lentivirus vector was purchased from Shanghai Genechem Co. LTD. (China). Centricon Plus-20 Centrifugal Filter was from Millipore (Billerica, MA). Cellomics Array Scan was from Thermo Scientific (Pittsburgh, PA). Fetal bovine serum, Trizol reagent, and Lipofectamine reagent were purchased from Invitrogen (Carlsbad, CA). M-MLV reverse transcriptase, dNTP, and RNase Inhibitor were from Promega (Madison, WI). DMEM and Opti-MEM were from Gibco-BRL (Grand Island, NY). TP800-Thermal Cycler Dice™ Real Time System was from Takara (Tokyo, Japan).
Generation of lentiviral particle
A scrambled shRNA (Scr shRNA) sequence, 5′-TTCTCCGAACGTGTCACGT-3′, was used as the non-silencing control. The shRNA sequence targeting IEF3B gene was 5′-GCATCTATGAAACTCCTTCTA-3′. The pGCSIL-GFP plasmid comprising EIF3B shRNA or Scr shRNA was cotransfected with the packaging plasmids (pHelper 1.0 and pHelper 2.0) into HEK293T cells using Lipofectamine 2000™ according to the manufacturer’s instruction. At 8 h after transfection, the cells were changed with fresh complete growth media and incubated for another 48 h. Then the culture media containing the viral particle was collected and centrifuged at 4,000g for 10 min to remove the cell debris and filtered through a 0.45-μm filter. The viral supernatant was further concentrated with a Centricon Plus-20 Centrifugal Filter at 4,000g for 10 min. The concentrated viral supernatant was aliquoted and kept at −80°C before use.
Lentivirus-mediated target gene knockdown
About 5 × 104 U87 cells were seeded onto the well of a six-well plate and incubated at 37°C with 5% CO2 until reaching 30% confluence. The concentrated viral supernatant was added into the culture medium at a multiplicity of infection (MOI) of 20. After 72 h, the cells were visualized under fluorescent microscope, which showed a transduction efficiency of about 90%.
Reverse transcription (RT)-PCR
Total mRNA samples of U87 cells or human glioblastoma cells from tissues obtained from ten patients were prepared with Trizol reagent according to the manufacturer’s instructions. Samples (2.0 μg) were used as templates to perform the RT-PCR assay. The RNA samples were mixed with 1 μl Oligo dT (0.5 μg/μl) and DEPC-H2O to a final volume of 9 μl. The mixture was incubated at 70°C for 10 min and immediately cooled on ice to allow the annealing of Oligo dT and template, followed by the addition of 4 μl 5 × RT buffer, 2 μl 10 mM dNTPs, 0.5 μl RNasin, 1 μl M-MLV-RTase and 3.5 μl DEPC H2O. The reaction system was incubated at 42°C for 1 h and at 70°C for 10 min to denature the M-MLV-RTase. The cDNA was kept at −80°C prior to use. For real-time PCR, the reaction system contained 10 μl SYBR Premix Ex Taq, 0.5 μl forward primer, 0.5 μl reverse primer, 1 μl cDNA, and 8.0 μl H2O. Primers used for real-time PCR were as follows: GAPDH forward: 5′-TGACTTCAACAGCGACACCCA-3′ and GAPDH reverse: 5′-AGGGGCCGGACTCGTCATACT-3′; EIF3B forward: 5′-CGGTGCCTTAGCGTTTGTG-3′ and EIF3B reverse: 5′-CGGTCCTTGTTGTTCTTCTGC-3′. Real-time RT-PCR was performed using a TP800 real-time PCR detection system with the following cycling conditions: (i) 15 s at 95°C and (ii) 45 cycles, with 1 cycle consisting of 5 s at 95°C and 30 s at 60°C. After real-time PCR amplification, the melting curve was monitored with the following conditions: 1 min at 95°C, 1 min at 55°C, followed by a temperature range from 55°C to 95°C increased by 0.5°C for every 4 s. GAPDH was employed as an internal reference under the same experimental conditions. Data were analyzed with 2−ΔΔCt method and the graph was plotted with GraphPad PRISM 4.0. The values were obtained through normalizing EIF3B copies to GAPDH copies.
Cell growth curve
Cells of the logarithmic phase were trypsinized, centrifuged, resuspended in the complete culture medium, and then seeded onto the 96-well plates (2,000 cells/well). The cells positive for GFP fluorescence were counted using a Cellomics Array Scan High Contents Screening Reader for 5 consecutive days. The growth curves were drawn accordingly.
BrdU assay
Analysis of DNA synthesis was performed with BrdU Cell Proliferation ELISA (#11647229001; Roche Diagnostics). Cells infected with EIF3B shRNA expressing lentivirus (Lenti-EIF3BshRNA) or scramble shRNA expressing lentivirus were seeded into 96-well plates (2,000 cells/well) and cultured for 24 or 96 h. BrdU diluted 100-fold in DMEM was added (10 μl/well) and incubated for 8 h. After treating with FixDenat for 30 min in the dark and with 10% BSA blocking solution, diluted anti-BrdU-POD antibody was added (100 μl/well) and incubated for 90 min at room temperature. Substrate solution (100 μl/well) was then added to develop color in dark for 15 min. Then, 10% H2SO4 (50 μl/well) was provided into each well and signals were measured at OD 490 nm.
Cell cycle analysis
Lentivirus-transduced U87 cells were cultured in a 6-cm dish until reaching 80% confluence, trypsinized, washed twice in PBS, and fixed with 70% pre-chilled ethanol at 4°C for 1 h. The fixed cells were washed and stained with propidium iodide (PI) mixture containing 50 µg/ml PI and 100 µg/ml ribonuclease in PBS for 45 min at 37°C. The cells were passed through a 300-mesh nylon net before the DNA content was determined by quantitative flow cytometry with standard optics of FACScan flow cytometer (Becton–Dickinson FACS Calibur). All the groups were performed in triplet and statistically analyzed.
FACS analysis of apoptosis
Analysis of apoptosis was performed with the Annexin V-APC Apoptosis Detection Kit according to the manufacturer’s instruction (#88-8007; eBioscience). Briefly, 48 h after infection, the adherent and nonadherent cells were collected, washed once in 1× binding buffer and resuspended in 1× staining buffer to 5 × 106 cells/ml. Then, 100 µl cells (about 5 × 105 cells) were incubated with 5 µl Annexin V-APC at room temperature for 15 min in the dark. Annexin V-stained cells were assessed using FACScan flow cytometer (Becton–Dickinson FACS Calibur).
Statistical analysis
All experiments were performed three times in triplicate. The data were analyzed with one-way ANOVA. Differences were considered statistically significant at P value <0.05.
Results
Expression of EIF3B in glioblastoma
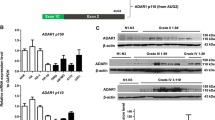
Ten glioblastoma tissue specimens from patients with different pathological status were analyzed for expression of EIF3B mRNA using real-time PCR assay. As shown in Fig. 1a, EIF3B expression in the specimens in Grade I, II, or III was not significantly different. Interestingly, one specimen in Grade IV showed higher EIF3B expression in comparison to the other nine specimens. However, more clinical samples were required to conclude whether there was a correlation between EIF3B expression and the pathological status of glioblastoma tissue.
Expression of EIF3B in human glioblastoma tissue and cell lines. a Data shown was obtained from fresh biopsy specimens from ten patients with glioblastoma (two Grade I, two Grade II, three Grade III, and three Grade IV lesions) and analyzed by real-time PCR assay. b RT-PCR analysis of EIF3B mRNA expression in five human glioma cell lines
Lentivirus-mediated knockdown of EIF3B gene in U87 glioblastoma cells
The expressions of EIF3B in five glioma cells were analyzed with RT-PCR (Fig. 1b). U-87, U-251, A172, and U373 cells have high EIF3B mRNA level. To study the function of EIF3B in glioma cells, U87 glioma cells were transduced with lentivirus expressing shRNA to knockdown EIF3B expression. The percentage of the GFP-expressing cells showed that about 90% of the cells were successfully transduced (Fig. 2a). As examined by real-time PCR (Fig. 2b), Lenti-EIF3BshRNA effectively downregulated the expression of EIF3B in U87 cells, compared to Lenti-Scr-shRNA (P < 0.001).
Lentivirus-mediated knockdown of EIF3B gene in U87 glioma cells led to inhibition of tumor proliferation in U87 cells. a Identification of knockdown efficiency of Lenti-EIF3B-shRNA. U87 cells were transduced with Lenti-Scr-shRNA or Lenti-EIF3B-shRNA as indicated in “Materials”. The total cell mRNA levels were analyzed with real-time PCR. P < 0.001, compared to control group (Scr shRNA). c U87 cells were transduced with Lenti-Scr-shRNA or Lenti-EIF3B-shRNA. The cells with a transduction rate more than 50% were trypsinized and seeded onto 96-well plates and then cultured for 5 consecutive days. The cells expressing green fluorescent protein were counted with the Cellomics Array Scan Reader and statistically analyzed. d Lenti-EIF3B-shRNA inhibited U87 cells proliferation by BrdU incorporation assay. U87 cells in the logarithmic phase were trypsinized and seeded onto 96-well plate. The cells were incubated at 37°C for 4 days and then OD 490 nm was measured. ***P < 0.001, compared to control group (Scr shRNA)
Knockdown of EIF3B inhibits tumor proliferation of U87 cells
After confirming the knockdown efficiency of Lenti-EIF3B-shRNA, lentiviral-transduced U87 cells were seeded onto 96-well plate and the cells positive for GFP fluorescence were counted with Cellomics Array Scan High Contents Screening Reader on each day for 5 consecutive days and the growth curves were drawn accordingly (Fig. 2c, P < 0.001). The result indicated that knockdown of EIF3B expression significantly suppressed the cell growth in U87 glioblastoma cells. To further study the impact of Lenti-EIF3B-shRNA on cell proliferation, BrdU incorporation assay was performed. As shown in Fig. 2d, Lenti-EIF3B-shRNA significantly inhibited the BrdU incorporation ability of U87 cells (P < 0.001), suggesting that EIF3B gene plays an important role in the DNA synthesis.
Knockdown of EIF3B induced the G0/G1 phase cell cycle arrest
To investigate whether the inhibitory effect of EIF3B on cell proliferation was associated with the cell cycle progression, lentiviral-transduced U87 cells were stained with PI solution and analyzed with FACS (Fig. 3a). Cell cycle analysis indicated that knockdown of EIF3B expression significantly increased the cell population in G0/G1 phase (P < 0.001) and reduced the cell population in the S-phase (Fig. 3b, P < 0.01), implying that EIF3B was involved in the cell cycle regulation of U87 glioblastoma cells. This result suggested that knockdown of EIF3B expression may inhibit cell growth through G0/G1 phase arrest.
Knockdown of EIF3B reduces the number of U87 cells in the S-phase. a Cell cycle distribution of lentivirus-transduced cells. U87 cells transduced with Lenti-Scr-shRNA or Lenti-EIF3B-shRNA were seeded onto the six-well plate. The cells were trypsinized at a confluency of 80%, stained with PI solution, and analyzed by FACS. The figure shows a representative result from three independent experiments. b Lenti-EIF3B-shRNA reduced the number of U87 cells in the S-phase. Statistical analysis showed that knockdown of EIF3B expression inhibited cell cycle progression through the S-phase. **P < 0.01 and ***P < 0.001, compared to control group (Scr shRNA)
Knockdown of EIF3B induces apoptosis of U87 cells
To study the function of EIF3B on U87 cell apoptosis, U87 cells were analyzed with FACS. Annexing-V staining result indicated that Lenti-EIF3B-shRNA significantly promoted cell apoptosis in U87 glioblastoma cells (Fig. 4, P < 0.05), underscoring that EIF3B may be involved in the cell survival pathway in U87 cells.
Knockdown of EIF3B promotes apoptosis of U87 cells. Lenti-Scr-shRNA or Lenti-EIF3B-shRNA transduced U87 cells were stained with Annexin V-APC and analyzed with FACS. The figure showed the results from three independent experiments. Lenti-EIF3B-shRNA promotes apoptosis in U87 cells. Statistical analysis showed that knockdown of EIF3B expression promotes U87 cell apoptosis. **P < 0.01, compared to control group (Scr shRNA)
Discussion
Malignant glioblastoma is one of the most lethal diseases in adulthood. The median survival of patients with Grade IV glioma, glioblastoma multiforme (GBM), is shorter than 15 months and the current first-line therapies for this devastating disease have only a palliative effect [9]. Glioblastoma cells are notorious for their rapid proliferation and widespread invasion to surrounding normal brain tissues [10]. The growth, proliferation, and migration are well organized by the harmonious expressions of abundant genes in the normal cells. Deregulations of this system resulting from extra- or intracellular disturbances initiate the tumorigenesis processes. Unraveling the mechanisms and indentifying target genes functioning in these processes are key steps for developing anti-cancer therapies. In the present study, we showed that EIF3B was expressed in fresh biopsy specimens of ten patients with glioblastoma and four glioblastoma cell lines. Knockdown of EIF3B level by EIF3B-shRNA expressing lentivirus subsequently inhibits the proliferation of U87 cells, arrests G0/G1-phase progression, and induces cell apoptosis. These results indicate EIF3B as a tumor-related gene in glioma cells, and suggest it as a target gene for anti-glioma therapy.
EIF3 is a multisubunit protein complex and plays an important role in several steps of the translation initiation pathway, which include recruiting the EIF2–GTP–MET–tRNA (iMet) ternary complex and other EIFs to the 40S ribosomal subunit to form the 43S preinitiation complex, mRNA recruitment, and subsequent scanning of the 5′ untranslated region (UTR) and start codon [11]. The HCV RNA genome contains an internal ribosome-entry site (IRES), which could specially bind with 40S ribosomal EIF3, allowing direct recognition of the start codon present in the 50-UTR of the viral mRNAs [12]. Among the EIF3 subunits, EIF3B is considered to be the major scaffolding subunit and it can bind directly to the IRES of HCV RNA [13]. Although EIF3B is considered to play an important role both in cellular and viral translation initiation, the cellular function of EIF3B is still unknown. Here, we demonstrated that U87 glioma cells had high level of EIF3B gene expression, and knockdown of EIF3B expression significantly inhibited the U87 cells growth. Cell cycle is closely related to cell proliferation and therefore the tumorigenesis processes [14]. This study indicated that downregulation of EIF3B expression significantly reduced S-phase cell number of U87 glioma cells and accumulated cells in the G0/G1 phase. This result implied that knockdown of EIF3B may be associated with an inhibition in DNA replication, which led to a reduced cell growth rate found in cell growth analysis (Fig. 2). An increase in the G0/G1 cell population may suggest that knockdown of EIF3B expression could promote the cells entering a senescence stage. However, the precise mechanism of how EIF3B regulates the cell cycle progression need to be further studied.
In conclusion, knockdown of EIF3B expression significantly arrests cell population in the G0/G1-phase and induces cell apoptosis, thereby suppressing cell proliferation and tumor growth of U87 glioma cells. Collectively, this study indicates that EIF3B plays a pivotal role in tumorigenesis of U87 glioma cells and may be a potential therapeutic target for anti-glioma therapy.
References
Haque A, Banik NL, Ray SK (2007) Emerging role of combination of all-trans retinoic acid and interferon-gamma as chemoimmunotherapy in the management of human glioblastoma. Neurochem Res 32:2203–2209
Pulkkanen KJ, Yla-Herttuala S (2005) Gene therapy for malignant glioma: current clinical status. Mol Ther 12:585–598
Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI et al (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98:7029–7036
Mayeur GL, Fraser CS, Peiretti F, Block KL, Hershey JW (2003) Characterization of eIF3k: a newly discovered subunit of mammalian translation initiation factor elF3. Eur J Biochem 270:4133–4139
Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW (2004) The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J Biol Chem 279:8946–8956
Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR et al (2008) Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA 105:18139–18144
Méthot N, Rom E, Olsen H, Sonenberg N (1997) The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J Biol Chem 272:1110–1116
ElAntak L, Tzakos AG, Locker N, Lukavsky PJ (2007) Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40S ribosomal subunit. J Biol Chem 282:8165–8174
Nakano I, Saya H (2010) Cancer stem cells in malignant glioma: the mechanism of cancer initiation and the therapeutic development. No Shinkei Geka 38:879–889
Melmed S (2006) Medical progress: Acromegaly. N Engl J Med 355:2558–2573
Hinnebusch AG (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31:553–562
Jackson RJ, Kaminski A (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985–1000
Pérard J, Rasia R, Medenbach J, Ayala I, Boisbouvier J, Drouet E et al (2009) Human initiation factor eIF3 subunit b interacts with HCV IRES RNA through its N-terminal RNA recognition motif. FEBS Lett 583:70–74
Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432:316–323
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, H., Ding, X., Zhou, C. et al. Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol Sci 33, 1057–1062 (2012). https://doi.org/10.1007/s10072-011-0894-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-011-0894-8