Abstract
Introduction
Celastrol is a promising therapeutic agent for the treatment of osteoarthritis (OA). However, the mechanism of action of celastrol is unclear. This study was aiming to identify the potential function of celastrol on OA and determine its underlying mechanism.
Method
Celastrol targets were collected from web database searches and literature review, while pathogenic OA targets were obtained from Online Mendelian Inheritance in Man (OMIM) and GeneCards databases. Transcriptomics data was sequenced using an Illumina HiSeq 4000 platform. Celastrol-OA overlapping genes were then identified followed by prediction of the potential function and signaling pathways associated with celastrol using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. A celastrol-target network was constructed to identify the candidate core targets of celastrol. The predictions were then validated by performing molecular docking and molecular dynamics simulation studies.
Results
In total, 96 genes were identified as the putative celastrol targets for treatment of OA. These genes were possibly involved in cell phenotype changes including response to lipopolysaccharide and oxidative stress as well as in cell apoptosis and aging. The genes also induced the mTOR pathway and AGE-RAGE signaling pathway at the intracellular level. Additionally, results indicated that 13 core targets including mTOR, TP53, MMP9, EGFR, CCND1, MAPK1, STAT3, VEGFA, CASP3, TNF, MYC, ESR1, and PTEN were likely direct targets of celastrol in OA. Finally, mTOR was determined as the most likely therapeutic target of celastrol in OA.
Conclusion
This study provides a basic understanding and novel insight into the potential mechanism of celastrol against OA.
Key Points • Our study provides a strong indication that further study of celastrol therapy in OA is required. • mTOR is the most likely therapeutic target of celastrol in OA. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is one of the most common diseases in middle-aged and elderly people. Despite its prevalence, there are no available disease-modifying osteoarthritis drugs (DMOADs) or other disease-modifying interventions due to lack of effective diagnostic tools for early detection of OA and monitoring disease progression [1]. Recently, there has been a rise in the number of researchers suggesting that there are different phenotypes or subpopulations of OA based on pathogenesis. Reports have also indicated that more than one pathogenetic mechanisms may be involved in the same patient during different phases of the disease [2]. Therefore, a single-target drug is not able to combat this complex multifactorial disease. A better therapeutic effect may be achieved by multi-targeted therapy through either drug combinations or a single drug simultaneously interacting with multiple targets.
Celastrol, an active ingredient isolated from Tripterygium wilfordii, has been used for centuries to treat inflammatory arthritis and autoimmune diseases in China. The molecule has multiple-target effects with reports indicating that its anti-inflammatory, antioxidant, and leptin sensitization effects have therapeutic effects in various inflammatory diseases [3], neurodegenerative diseases [4], and metabolic disorders [5]. Celastrol’s therapeutic effects suggest that it could play a role in OA associated with synovial inflammation and cartilage breakdown. Although some reports on the positive effects of celastrol on OA exist [6, 7], the exact mechanisms of anti-osteoarthritis of celastrol are still largely unknown. Therefore, there is need to conduct further studies to identify the targets interacting with celastrol and elucidate its underlying mechanism in OA treatment.
Currently, mechanistic studies on celastrol mainly follow the conventional single target one disease paradigm which limits its evaluation. This is because the therapeutic effect of multi-target drugs is usually attributed to the synergistic action between various targets involved in the different progression of the disease. In recent years, new experimental technologies are generating numerous multi-omics data which is providing information on complex diseases including OA thus increasing the number of potential drug targets [8]. However, drug development has not matched the pace of these experimental technologies due to the fact that the disease network does not provide information on drug-disease associations which is the main purpose of this study.
Network pharmacology is a novel pharmacological approach used to access the interactions of specific nodes or modules within integrated drug targets and biological networks [9, 10]. Unlike traditional pharmacological approaches, it provides descriptions of drug action at the molecular level from a biological balance perspective. Therefore, it is considered to be a promising discipline for evaluation of complex diseases and multi-target drugs. In this study, we combined the transcriptome, network pharmacology, and computer-aided drug design methods using an integrated analysis strategy to explore the underlying mechanism of the action of celastrol on OA. Results from this study may provide new perspectives in understanding the potential mechanisms of celastrol in treating OA and drug repositioning.
Materials and methods
Collection of celastrol targets through web database and literature review
Prediction of celastrol targets using web databases was based on its chemical similarity with known ligands. The chemical structure of celastrol was first retrieved from the PubChem database (http://www.pubchem.ncbi.nlm.nih.gov) [11] followed by prediction of possible targets using the web-available SwissTargetPrediction database (http://www.swisstargetprediction.ch). Similarity ensemble approach (SEArch, http://sea.bkslab.org/) was then performed [12, 13] followed by selection of putative targets with threshold values of >0 probability or P <0.05.
Celastrol targets were supplemented using the Traditional Chinese medicine systems pharmacology database (TCMSP, http://lsp.nwsuaf.edu.cn/tcmsp.php) [14] followed by a comprehensive literature survey to retrieve known targets [15,16,17]. Finally, all the putative targets were standardized using UniProt database (https://www.uniprot.org/) and then filtered using the term “Homo sapiens” [18].
Identification of celastrol target mRNA transcripts using transcriptome analysis
Human primary chondrocytes were harvested from adult human articular cartilage using collagenase digestion method. Cartilage tissues were obtained from surgical specimens of OA patients with KL grades 3–4 requiring total knee replacement. All patients signed written informed consents before surgery with all the procedures described being approved by the Ethics Committee of First Affiliated Hospital of Harbin Medical University (IRB: 2018139).
Collected chondrocytes were then cultured in sterile T25 culture flasks and maintained in DMEM/high glucose (Hyclone Laboratories) media supplemented with 10% fetal bovine serum (Gibco, Australia Origin) and 1% Penicillin-Streptomycin solution. The culture flasks were then placed in a CO2 incubator at 37°C. After growing to 80% confluence, the 2nd passage OA chondrocytes were treated with 200-nM celastrol (purity≥98%, Sigma-Aldrich, Germany) or the vehicle control medium. The concentration of celastrol in this study was determined by reference to published data and our preliminary experimental results [6, 7].
Total RNA from chondrocytes was extracted after 24-h treatment using TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the protocol described by the manufacturer [19]. Subsequently, 3μg of high-quality RNA samples (defined by 260/280 ratios of >1.9 and RNA integrity number scores of >8) was used to generate RNA libraries using rRNA-depleted RNA by NEBNext® Ultra™ Directional RNA Library Prep Kit from Illumina (NEB, USA). Transcriptome sequencing was conducted at the Novogene Bioinformatics Institute (Beijing, China) using an Illumina HiSeq 4000 platform. Clean reads (after removing adapter or poly-N and low-quality reads) were used for the downstream analysis.
Collection of pathogenic OA targets
Targets related to OA were obtained from Online Mendelian Inheritance in Man (OMIM, www.ncbi.nlm.nih.gov/omim) and GeneCards databases (www.genecards.org) using the term “osteoarthritis” in the search engine [20, 21]. Targets described in OMIM gene category in the OMIM database and targets with GeneCards score higher than the median in the GeneCards databases were selected for displaying the results. All selected targets were filtered with the background organism “Homo sapiens” and mapped on the UniProt database to obtain the gene symbol for further homology analysis. All duplicates and unmapped targets were removed from the data set.
Enrichment analysis of celastrol targets
Functional annotation and pathways enrichment analysis of celastrol-OA overlapping genes was performed by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using R/Bioconductor clusterProfiler (version 3.12.0). Enriched GO terms were classified as molecular function (MF), cellular component (CC), and biological process (BP).
Construction of celastrol-OA targets interaction network and screening of core targets
To calculate the combined score between each protein-protein pair, celastrol-OA common target genes were imported into STRING database (https://string-db.org/, version 11.0) [22]. Interactions with confidence scores ranging from 0.4 to 1 were selected and uploaded into the Cytoscape software to construct protein-protein interaction (PPI) networks. The topological parameters of the network were analyzed using a NetworkAnalyzer tool in Cytoscape. The hub nodes, which were defined as having degree, betweenness, and closeness scores greater than the median, were classified as the candidate core targets of celastrol [23].
Validation of celastrol-target interactions using molecular docking
Validation of the celastrol-target associations was done by molecular docking simulation of each putative target interacting with celastrol [24]. The naïve 3D structure of celastrol and protein complex was retrieved from PubChem database and RCSB Protein Data Bank (https://www.rcsb.org/) [25], respectively, with extraction of the structures being done using AutoDock Tools (v1.5.6). Before docking, processed Protein Data Bank (PDB) files were converted to PDBQT format by adding polar hydrogens and assigning charges. All docking calculations were performed by the AutoDock Vina software (v1.1.2), while the semi-flexible docking method with semi-empirical energy functions was used to interpret docking results. The lowest energy structure in the docking results was taken as the best-docked conformation followed by visualization using PyMOL (v2.3.0).
Molecular dynamics (MD) simulation
When compared to molecular docking, molecular dynamics (MD) simulation is an important and widely used tool which provides more realistic and reliable results to assess the stability of drug-target complexes. Therefore, MD simulation was performed in GROMACS release 2019.4 package with GROMOS96 54a7 force field to further verify the potential interaction identified above using network pharmacology and molecular docking methods. The topology of celastrol was first generated using PRODRG2 server followed by embedding of the target-celastrol system in a cubic box with appropriate size to maintain periodic boundary conditions and the simple point charge water solvation model (112662 water molecules). The overall system was neutralized by adding 3 Na+ ions in the solution. SHAKE algorithm was used to constrain all bond lengths involving hydrogen atoms, while long-range electrostatic interactions were treated using the Particle Mesh Ewald method with a cutoff of 12 Å. NPT and NVT ensemble equilibration steps were done after the system was suitably minimized. Final productions run for each system was done using Parrinello-Rahman barostat to maintain a constant temperature and pressure during the simulation time. A 2-fs time step was applied with the production MD simulation of the protein-ligand complex being done for 50 ns.
Statistical analysis
In this study, the differential gene expression was implemented by the edgeR statistical software. Genes with absolute fold change ≥2 and P<0.05 were considered as differentially expressed genes (DEGs) and selected as celastrol targets. For GO and KEGG enrichment analysis, R/Bioconductor clusterProfiler (version 3.12.0) was used. Terms with adjusted P value less than 0.05 were considered significantly enriched.
Results
There were overlapping networks of celastrol target and OA-related genes
In total, 1325 human genes associated with OA were collected from OMIM and GeneCards databases. A total of 453 candidate genes (after removing the duplicate genes) targeted by celastrol were collected with 44 putative targets being predicted by TCMSP, SEArch, and Swiss Target Prediction database, 50 verified targets obtained from literature review, and 373 available after transcriptome analysis. DEGs between celastrol treatment and OA after transcriptome analysis are shown in Fig. 1, while all the RNA sequencing data has been deposited at Sequence Read Archive (SRA) database under accession ID PRJNA602231. In the two data sets, there are 96 overlapping genes (Fig. 3a) which could be potential drug targets for the treatment of OA.
Transcriptomic analysis of differential mRNA expression in human OA chondrocytes cultured with or without celastrol treatment (200nM, 24h). a Volcanic plot of the differential mRNA expression analysis. The differential expression was assessed using RNA-seq, n=3. Red and blue circles represent mRNA downregulation and upregulation, respectively. Green circles indicate mRNAs with P value >0.05. b Heatmap of differentially expressed genes
Celastrol affects biological functions
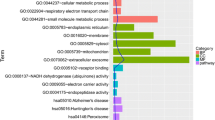
GO and KEGG enrichment analyses were performed to study the biological functions resulting from exposure to celastrol. A total number of 932 enriched GO BPs (Supplementary Table 1), 57 enriched GO CCs (Supplementary Table 2), 93 enriched GO MFs (Supplementary Table 3), and 127 KEGG signaling pathways (Supplementary Table 4) were identified. Common genes of celastrol-OA targets were categorized by their functions mainly including response to lipopolysaccharide (LPS), metabolic process, cell apoptosis and aging, inflammasome complex, insulin receptor substrate binding, and ubiquitin-like protein ligase binding (Fig. 2a). In addition, the KEGG term annotation analysis for the genes in response to celastrol was involved in several pathways including apoptosis protein processing, cellular senescence, autophagy, mTOR signaling pathway, Wnt signaling pathway, AGE-RAGE signaling pathway in diabetic complications, non-alcoholic fatty liver disease, fluid shear stress and atherosclerosis, and Alzheimer’s disease (Fig. 2b).
Gene ontology (GO) functional enrichment and KEGG pathway enrichment analysis of common genes of celastrol-OA targets. a Bar chart of predominant GO terms. The horizontal axis represents the enrichment scores for each GO term, while the vertical axis represents the GO annotations. Three different colors represent distinct terms of biology process (BP), cellular component (CC), and molecular function (MF). b Bubble plot representative of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment results. The horizontal axis represents the ratio between the number of genes related to the term in the pathway and the total number of genes, while the vertical axis represents the name of the pathway. Size of the bubble represents the number of genes, and the color of bubble represents the adjusted P values
Identifying core celastrol targeted proteins
The 96 common genes described above were used to construct a PPI network of 93 codes and 1333 edges (Fig. 3b). Screening of candidate core targets was done with conditions maintained at a degree of >26, closeness >0.56790, and betweenness >0.002344. Detailed topological parameters of the candidate core targets are presented in Supplementary Table 5. Fifteen nodes with gene names were identified as core targets using the two-fold median value of node degree as a threshold. They include TP53, TNF, VEGFA, CASP3, JUN, MAPK1, STAT3, MYC, MMP9, FN1, CCND1, ESR1, EGFR, mTOR, and PTEN (Table 1). The genes had a higher degree than other genes in the PPI network. This means that during the drug action, these targets participate in more interactions than other cellular components [10].
Overlapping genes of celastrol-OA targets and protein-protein interaction (PPI) networks. a Orange circles represent 96 overlapping genes that are common to celastrol targets and OA likely pathogenic genes. b PPI network containing 96 nodes and 1333 edges. The circles represent individual PPI network nodes with gene names. The color intensity of the nodes together with the node size is proportional to the degree value
Molecular docking model of celastrol binding to its core targets
Assessment of a good drug target should not only focus on the importance of the target in the biological process but also on the accessibility of the target to the drug. With this in mind, we performed molecular docking to further investigate the binding mechanisms of celastrol and 15 of the core targets. The docking information of celastrol (as ligand) against individual target proteins is displayed in Table 2. The results indicate that celastrol could bind to 13 of the 15 candidate core targets (binding energy < 0 kJ·mol−1) suggesting that these proteins are putative direct targets of celastrol in OA. However, the accuracy of the results must be first confirmed using PPI predict data. In addition, celastrol interacted strongly with 10 of the core targets including mTOR (PDB: 4JSX), TP53 (PDB: 4XR8), MMP9 (PDB: 2OVX), EGFR (PDB: 5EDR), cyclin-D1 (PDB: 2W96), MAPK1 (PDB: 5NHV), STAT3 (PDB: 6NUQ), VEGFA (PDB: 3QTK), caspase-3 (PDB: 4JJE), and tumor necrosis factor (PDB: 3IT8). Evidence of a strong interaction was given by the binding energy being less than −5.0 kJ·mol−1.
Four representative docking models with the strongest binding capacity(top 4 lowest binding energy)were visualized using PyMOL with results showing that celastrol could access the active pockets of all the selected proteins (Fig. 4). Hydrogen bonding and hydrophobic interactions are the main binding modes between the pairs of celastrol and mTOR and TP53, while celastrol binds to MMP9 and EGFR mainly through hydrophobic contacts. In addition, there were three hydrogen bonds between celastrol and amino acid residues of mTOR explaining the high binding energy of mTOR-celastrol complex. These results indicated that celastrol has a higher affinity for mTOR than other candidate targets suggesting that mTOR should be the most likely therapeutic target of celastrol in OA for effective therapy.
Molecular docking models showing celastrol binding to its targets. a Serine/threonine-protein kinase mTOR (mTOR). b Cellular tumor antigen p53 (TP53). c Matrix metalloproteinase-9 (MMP9). d Epidermal growth factor receptor (EGFR). Celastrol molecule is shown as sticks, while the hydrogen bonds between the celastrol and the active site amino acid residues are shown as blue solid lines. The yellow dashed lines show salt bond, and the gray dashed lines show hydrophobic interactions
MD simulation confirmed results of mTOR-celastrol complex binding
Based on the docking results, a 50-ns MD simulation of the mTOR (PDB: 4JSX, with the lowest binding energy)-celastrol complex at a temperature of 300K and a pressure of 1atm was done. The Docking pose extracted at 0 ns, and the interacting amino acid residues for the mTOR-celastrol complex are shown in Fig. 5b. The root-mean-square deviation (RMSD) value, an important indicator of the stability of the complex, indicated that the system reached equilibrium at 3 ns and remained highly stable throughout the simulation time of 50 ns with less than 0.2 Å fluctuations (Fig. 5a). MD simulation results confirm the results obtained after molecular docking.
Molecular dynamics results of mTOR (PDB: 4JSX)-celastrol. a Root-mean-square deviation (RMSD) plot of the backbone atoms of the mTOR-celastrol during the 50-ns MD simulation. b Docking pose of celastrol with mTOR extracted at 0 ns. Celastrol molecule is shown as sticks with cyan color. The active site residues are shown as orange sticks, while the yellow dashed lines show the hydrogen bonds between the celastrol and the active site amino acid residues
Discussion
A clear understanding of the pathogenesis of OA has ensured that it is no longer considered a natural and inevitable consequence of normal aging but rather a treatable disorder with research focusing on the search for effective treatments. Celastrol is a promising therapeutic drug for OA. However, the underlying mechanisms of anti-osteoarthritis after celastrol treatment remain unclear. The aim of this study was to understand the potential pharmacological mechanism of celastrol against OA.
In this study, we combined the information collected from publicly available databases and our own transcriptomics data of celastrol treatment for OA and used a network pharmacology approach to reveal the network characteristics of celastrol and to explore the drug targets. Through this research, 96 genes were identified as potential candidates for celastrol treatment of OA. The genes were subsequently analyzed for enrichment of GO annotations and KEGG pathways to determine the underlying mechanism of celastrol action. Results of GO BPs analysis show that celastrol may play a key role in the altered cell phenotype including response to LPS, oxidative stress, metabolic process, cell apoptosis and aging, as well as extracellular matrix organization. Among them, celastrol regulating apoptosis and response to LPS-induced cell damage are in line with previous findings of celastrol treatment of OA [26, 27]. Although the regulation of cell aging, oxidative stress, and metabolic process by celastrol has not yet been reported in OA, its ability to induce these cell phenotype changes has been observed in other diseases [28,29,30]. It is also interesting to note the potential role of celastrol in regulating the aging of cells, since the causal role of senescent cells in the pathogenesis of OA has recently been confirmed [31]. Results obtained from KEGG pathway analysis provide a novel insight into the molecular mechanisms of celastrol’s action against OA. It depicted various signaling pathways such as mTOR signaling pathway, Wnt signaling pathway, and AGE-RAGE signaling pathway were induced by celastrol. These predicting pharmacological mechanisms have been proven to be related to the pathogenesis of OA, which provides a strong indication that further study of celastrol therapy in OA is required. Interestingly, the common targets for celastrol treatment of OA were also enriched in pathological changes associated with diabetes, non-alcoholic fatty liver disease (NAFLD), atherosclerosis, and Alzheimer’s disease. As these diseases share the same risk factors with OA and the potential of celastrol as a pharmacological intervention against obesity, NAFLD, atherosclerosis, and Alzheimer’s disease has been established in the recent studies [32, 33], it can be reasonably concluded that celastrol may be applicable to OA patients with these comorbid diseases. After PPI analysis and molecular docking, mTOR was identified as one of the core and direct targets for celastrol in OA, which agrees with previous studies. These studies have shown the ability of celastrol to regulate the gene or protein expression of mTOR [34, 35]. Furthermore, we previously reported that mTOR is the key player for longevity and aging and chondrocyte survival and the master negative regulator of autophagy [36, 37]. Therefore, it is not surprising that mTOR could serve as a potential therapeutic target for improving OA outcome [38], so our finding again demonstrated that celastrol might serve as a highly promising therapeutic agent for treatment of OA.
In addition, our study identified TP53 as a potential functional target for celastrol therapy, which is consistent with the findings previously published. In the previous studies, Lee et al. described the role of celastrol on the activation of P53 [39]. Zhang et al. reported celastrol may contribute to induction of cell apoptosis in APL through its role in P53 activation [40]. In the research of OA, P53 has been clearly shown to be related to chondrocyte apoptosis [41]. In addition, an inhibitor of p53/MDM2 interaction, UBX0101, is reported to attenuate the development of post-traumatic OA through selective elimination of senescent cells in osteoarthritic condition [42]. In our GO analysis, the function that celastrol modulates cell aging is attributed partly to this P53. However, as far as we know, there has been no report about the interaction between celastrol and this target in OA. Therefore, the identification of TP53 target might provide a new possible explanation for celastrol’s efficiency in the treatment of OA.
Lastly, different from our results, in the previous studies of OA, the pharmacological effects of celastrol were mostly attributed to its regulation of NF-κB signaling pathway. NF-κB is a key transcription factor regulating inflammatory responses [43]. Therefore, these previous findings reflected the anti-inflammatory effect of this drug. Among the core targets we screened, STAT3 and TNF also play a critical role in inflammation. However, besides regulating inflammatory reaction, our results also indicate that celastrol may exert its anti-osteoarthritis effect through various mechanisms, including regulating apoptosis, autophagy, and cell senescence. In fact, anti-inflammation drugs such as TNF inhibitor have been proved to fail to modify the course of OA in the clinical trials. OA is also considered to be the result of multiple targets and cellular interaction, not simply a consequence of inflammation, which just fits the network concept of network pharmacology.
In closing, this study applied the combination of transcriptomics, network pharmacology, and computer-aided drug design methods to uncover the underlying mechanism of celastrol. Approaches described in the study may be useful in investigating how multi-target drugs act on complex diseases. Application of this strategy will result in the emergence of a number of novel therapeutics for OA. Although additional experiments are necessary to validate these findings, the data presented in this study provides a basic understanding of therapeutic effects and potential mechanism of celastrol against OA.
Data availability
The associated raw sequencing data in this study has been deposited in the Sequence Read Archive (SRA) under accession ID PRJNA602231. All other data generated or analyzed during this study are included within the article and its supplementary files.
Abbreviations
- BP:
-
Biological process
- CC:
-
Cellular component
- DMOADs:
-
Disease-modifying osteoarthritis drugs
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MD:
-
Molecular dynamics
- MF:
-
Molecular function
- OMIM:
-
Online Mendelian Inheritance in Man
- OA:
-
Osteoarthritis
- PPI:
-
Protein-protein interaction
- PDB:
-
Protein Data Bank
- RMSD:
-
Root-mean-square deviation
- SRA:
-
Sequence Read Archive
- SEArch:
-
Similarity ensemble approach
- TCMSP:
-
Traditional Chinese medicine systems pharmacology
References
Oo WM, Yu SP, Daniel MS, Hunter DJ (2018) Disease-modifying drugs in osteoarthritis: current understanding and future therapeutics. Expert Opin Emerg Drugs 23(4):331–347. https://doi.org/10.1080/14728214.2018.1547706
Karsdal MA, Michaelis M, Ladel C, Siebuhr AS, Bihlet AR, Andersen JR, Guehring H, Christiansen C, Bay-Jensen AC, Kraus VB (2016) Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future. Osteoarthr Cartil 24(12):2013–2021. https://doi.org/10.1016/j.joca.2016.07.017
Venkatesha SH, Dudics S, Astry B, Moudgil KD (2016) Control of autoimmune inflammation by celastrol, a natural triterpenoid. Pathog Dis 74(6):ftw059. https://doi.org/10.1093/femspd/ftw059
Li J, Hao J (2019) Treatment of neurodegenerative diseases with bioactive components of Tripterygium wilfordii. Am J Chin Med 47(4):769–785. https://doi.org/10.1142/s0192415x1950040x
Liu J, Lee J, Salazar Hernandez MA, Mazitschek R, Ozcan U (2015) Treatment of obesity with celastrol. Cell 161(5):999–1011. https://doi.org/10.1016/j.cell.2015.05.011
Wang W, Ha C, Lin T, Wang D, Wang Y, Gong M (2018) Celastrol attenuates pain and cartilage damage via SDF-1/CXCR4 signalling pathway in osteoarthritis rats. J Pharm Pharmacol 70(1):81–88. https://doi.org/10.1111/jphp.12835
Ding QH, Cheng Y, Chen WP, Zhong HM, Wang XH (2013) Celastrol, an inhibitor of heat shock protein 90beta potently suppresses the expression of matrix metalloproteinases, inducible nitric oxide synthase and cyclooxygenase-2 in primary human osteoarthritic chondrocytes. Eur J Pharmacol 708(1-3):1–7. https://doi.org/10.1016/j.ejphar.2013.01.057
Ruiz-Romero C, Rego-Perez I, Blanco FJ (2018) What did we learn from ‘omics’ studies in osteoarthritis. Curr Opin Rheumatol 30(1):114–120. https://doi.org/10.1097/bor.0000000000000460
Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4(11):682–690. https://doi.org/10.1038/nchembio.118
Berger SI, Iyengar R (2009) Network analyses in systems pharmacology. Bioinformatics 25(19):2466–2472. https://doi.org/10.1093/bioinformatics/btp465
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 37(Web Server issue):W623–W633. https://doi.org/10.1093/nar/gkp456
Gfeller D, Michielin O, Zoete V (2013) Shaping the interaction landscape of bioactive molecules. Bioinformatics 29(23):3073–3079. https://doi.org/10.1093/bioinformatics/btt540
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK (2007) Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25(2):197–206. https://doi.org/10.1038/nbt1284
Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L (2014) TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 6:13. https://doi.org/10.1186/1758-2946-6-13
Cascao R, Fonseca JE, Moita LF (2017) Celastrol: a spectrum of treatment opportunities in chronic diseases. Front Med (Lausanne) 4:69. https://doi.org/10.3389/fmed.2017.00069
Chen SR, Dai Y, Zhao J, Lin L, Wang Y, Wang Y (2018) A mechanistic overview of triptolide and celastrol, natural products from Tripterygium wilfordii Hook F. Front Pharmacol 9:104. https://doi.org/10.3389/fphar.2018.00104
Hu M, Luo Q, Alitongbieke G, Chong S, Xu C, Xie L, Chen X, Zhang D, Zhou Y, Wang Z (2017) Celastrol-induced Nur77 interaction with TRAF2 alleviates inflammation by promoting mitochondrial ubiquitination and autophagy. Mol Cell 66(1):141–153.e146. https://doi.org/10.1016/j.molcel.2017.03.008
The UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47(D1):D506–d515. https://doi.org/10.1093/nar/gky1049
Zhang J, Li C, Zheng Y, Lin Z, Zhang Y, Zhang Z (2017) Inhibition of angiogenesis by arsenic trioxide via TSP-1-TGF-β1-CTGF-VEGF functional module in rheumatoid arthritis. Oncotarget 8(43):73529–73546. https://doi.org/10.18632/oncotarget.19867
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 54:1.30.31–31.30.33. https://doi.org/10.1002/cpbi.5
Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A (2015) OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43(Database issue):D789–D798. https://doi.org/10.1093/nar/gku1205
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–d613. https://doi.org/10.1093/nar/gky1131
Nacher JC, Schwartz JM (2008) A global view of drug-therapy interactions. BMC Pharmacol 8:5. https://doi.org/10.1186/1471-2210-8-5
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3(11):935–949. https://doi.org/10.1038/nrd1549
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Liu DD, Zhang BL, Yang JB, Zhou K (2020) Celastrol ameliorates endoplasmic stress-mediated apoptosis of osteoarthritis via regulating ATF-6/CHOP signalling pathway. J Pharm Pharmacol 72(6):826–835. https://doi.org/10.1111/jphp.13250
Li X, Wei W, Zhao Z, Lv S (2019) Tripterine up-regulates miR-223 to alleviate lipopolysaccharide-induced damage in murine chondrogenic ATDC5 cells. Int J Immunopathol Pharmacol 33:2058738418824521. https://doi.org/10.1177/2058738418824521
Du Z, Zhang W, Wang S, Zhang J, He J, Wang Y, Dong Y, Huo M (2019) Celastrol protects human retinal pigment epithelial cells against hydrogen peroxide mediated oxidative stress, autophagy, and apoptosis through sirtuin 3 signal pathway. J Cell Biochem 120(6):10413–10420. https://doi.org/10.1002/jcb.28326
Xu XJ, Zhao WB, Feng SB, Sun C, Chen Q, Ni B, Hu HY (2017) Celastrol alleviates angiotensin II-mediated vascular smooth muscle cell senescence via induction of autophagy. Mol Med Rep 16(5):7657–7664. https://doi.org/10.3892/mmr.2017.7533
Li L, Wang B, Li Y, Li L, Dai Y, Lv G, Wu P, Li P (2020) Celastrol regulates bone marrow mesenchymal stem cell fate and bone-fat balance in osteoporosis and skeletal aging by inducing PGC-1α signaling. Aging (Albany NY) 12(17):16887–16898. https://doi.org/10.18632/aging.103590
Sherwood J (2019) Osteoarthritis year in review 2018: biology. Osteoarthr Cartil 27(3):365–370. https://doi.org/10.1016/j.joca.2018.10.005
Zhang Y, Geng C, Liu X, Li M, Gao M, Liu X, Fang F, Chang Y (2017) Celastrol ameliorates liver metabolic damage caused by a high-fat diet through Sirt1. Mol Metab 6(1):138–147. https://doi.org/10.1016/j.molmet.2016.11.002
Allen SD, Liu YG, Kim T, Bobbala S, Yi S, Zhang X, Choi J, Scott EA (2019) Celastrol-loaded PEG-b-PPS nanocarriers as an anti-inflammatory treatment for atherosclerosis. Biomater Sci 7(2):657–668. https://doi.org/10.1039/c8bm01224e
Nie Y, Fu C, Zhang H, Zhang M, Xie H, Tong X, Li Y, Hou Z, Fan X, Yan M (2020) Celastrol slows the progression of early diabetic nephropathy in rats via the PI3K/AKT pathway. BMC Complement Med Ther 20(1):321. https://doi.org/10.1186/s12906-020-03050-y
Li X, Zhu G, Yao X, Wang N, Hu R, Kong Q, Zhou D, Long L, Cai J, Zhou W (2018) Celastrol induces ubiquitin-dependent degradation of mTOR in breast cancer cells. Onco Targets Ther 11:8977–8985. https://doi.org/10.2147/ott.S187315
Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, Lussier B, Roughley P, Lagares D, Pelletier JP, Martel-Pelletier J, Kapoor M (2015) Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis 74(7):1432–1440. https://doi.org/10.1136/annrheumdis-2013-204599
Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F (2003) Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426(6967):620. https://doi.org/10.1038/426620a
Pal B, Endisha H, Zhang Y, Kapoor M (2015) mTOR: a potential therapeutic target in osteoarthritis? Drugs R D 15(1):27–36. https://doi.org/10.1007/s40268-015-0082-z
Lee JH, Choi KJ, Seo WD, Jang SY, Kim M, Lee BW, Kim JY, Kang S, Park KH, Lee YS, Bae S (2011) Enhancement of radiation sensitivity in lung cancer cells by celastrol is mediated by inhibition of Hsp90. Int J Mol Med 27(3):441–446. https://doi.org/10.3892/ijmm.2011.601
Zhang X, Yang J, Chen M, Li L, Huan F, Li A, Liu Y, Xia Y, Duan JA, Ma S (2016) Metabolomics profiles delineate uridine deficiency contributes to mitochondria-mediated apoptosis induced by celastrol in human acute promyelocytic leukemia cells. Oncotarget 7(29):46557–46572. https://doi.org/10.18632/oncotarget.10286
Ito K, Maruyama Z, Sakai A, Izumi S, Moriishi T, Yoshida CA, Miyazaki T, Komori H, Takada K, Kawaguchi H, Komori T (2014) Overexpression of Cdk6 and Ccnd1 in chondrocytes inhibited chondrocyte maturation and caused p53-dependent apoptosis without enhancing proliferation. Oncogene 33(14):1862–1871. https://doi.org/10.1038/onc.2013.130
Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, Chung JW, Kim DH, Poon Y, David N, Baker DJ, van Deursen JM, Campisi J, Elisseeff JH (2017) Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med 23(6):775–781. https://doi.org/10.1038/nm.4324
Jimi E, Fei H, Nakatomi C (2019) NF-κB signaling regulates physiological and pathological chondrogenesis. Int J Mol Sci 20(24). https://doi.org/10.3390/ijms20246275
Code availability
Not applicable.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81771748) and Harbin Technology Bureau Research Fund (2017RAQXJ196).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
Ethical approval was obtained from the First Affiliated Hospital of Harbin Medical University.
Consent to participate
Written informed was acquired from each participant before surgery.
Consent for publication
Informed consent for publication was obtained from each study participant and each patient.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dai, S., Wang, H., Wang, M. et al. Comparative transcriptomics and network pharmacology analysis to identify the potential mechanism of celastrol against osteoarthritis. Clin Rheumatol 40, 4259–4268 (2021). https://doi.org/10.1007/s10067-021-05726-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05726-3