Abstract
Objectives
This study evaluated expression of circRNA in primary Sjögren’s syndrome (pSS) patients so as to find novel biomarkers for pSS screening and discussed possible role of circRNA in pSS. We also evaluated expression profile of circRNA in systemic lupus erythematosus (SLE) patients.
Methods
Microarray analysis detected circRNA expression in PBMCs from five paired pSS, SLE patients, and controls. Then, differentially expressed circRNAs were validated in 30 pSS patients as compared to 30 SLE patients, healthy controls. CircRNAs interacting with miRNAs were discussed by Arraystar’s homemade miRNA target prediction software. ROC analysis assessed the diagnostic value.
Results
We identified 234 differentially expressed circRNAs in pSS patients and verified five selected circRNAs (including hsa_circRNA_001264, hsa_circRNA_104121, hsa_circRNA_045355, hsa_circRNA_103461, hsa_circRNA_105034). Expression of hsa_circRNA_001264, hsa_circRNA_104121, and hsa_circRNA_045355 was strongly related to some clinical, laboratory parameters, and disease activity index in pSS patients. ROC analysis indicated potential diagnostic ability for the three circRNAs in pSS patients. One hundred and forty-eight circRNAs were differently expressed between lupus patients and controls.
Conclusion
This study provides evidence that hsa_circRNA_001264, hsa_circRNA_104121, and hsa_circRNA_045355 might be biomarkers for pSS, correlate with pSS etiology.
Key Points | |
• Many circRNAs were dysregulated in pSS patients. | |
• Differentially expressed circRNAs correlated with pSS clinical, laboratory features. | |
• CircRNAs may be biomarkers for pSS. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary Sjögren’s syndrome (pSS) is an inflammatory disorder, mainly destructs glands in mouth and eyes. These kinds of patients sometimes appeared extra-glandular characteristics, for instance, tubulointerstitial nephritis, and autoimmune hepatitis. These manifestations are mainly mediated through the overexpression of autoantibodies, which may result in hypergammaglobulinemia. Although the etiology of pSS is still unclear, genetics, environmental factors may have contributed significantly in the disease.
Balance of immunity in human body is effective to defend external infection or damage, whereas dysregulated immunity can lead to aggravated inflammatory response [1]; disease occurrence, such as cardiovascular disease; and rheumatoid arthritis (RA) [2, 3]. Investigations showed that microRNAs perform significantly in immune response [4, 5]. Recently, circular RNAs (circRNAs) are accepted importantly for regulating inflammatory response and affect biological function of immune cells. CircRNAs form covalently closed continuous loop structures [6]. There are numerous binding sites for circRNAs. They can bind to miRNAs and therefore act through absorption of miRNAs and affect some other RNAs by base pairing [7, 8]. After binding to different downstream proteins, circRNAs suppress activity of the proteins [9].
To date, reports began to elucidate the aberrantly expressed circRNAs in inflammatory autoimmune disorders, including RA and lupus [10,11,12]. Some circRNAs have been demonstrated relation to the pathogenesis of the disorders [12, 13]. Thus, searching for circRNAs that may be considered as biomarkers for pSS may improve to understand the complex pathogenesis of pSS. In the present study, we firstly tested expression profile of circRNA within pSS patients, then validated the aberrantly expressed circRNAs. The potential of circRNAs as the biomarkers for pSS was assessed as well. Moreover, we discussed circRNA in systemic lupus erythematosus (SLE) patients compared with previous studies.
Methods
Participants
Thirty-five Chinese Han pSS patients (32 women, 3 men; age 40.5 ± 10.5 years) were recruited. pSS patients were admitted according to 2002 American-European Consensus Group Classification Criteria [14]. Thirty-five SLE patients (33 women, 2 men; age 38.6 ± 11.5 years) were selected, and we recruited 35 healthy volunteers (32 women, 3 men; age 37.9 ± 9.8 years). Diagnosis of SLE was compliable with American College of Rheumatology (ACR) criteria [15]. All the patients were treatment naïve. The present study was conducted by two stages: discovery and validation set. First, five pSS, SLE and five volunteers were used, then an independent validation set including 30 pSS, SLE, 30 volunteers was recruited. This study was admitted by Southwest Medical University. Written consent was collected from individual participant.
Assessment of pSS activity and serological parameters
Demographic, clinical data were obtained from the participants (Table 1). Disease activity of pSS expressed as Sjögren’s Syndrome Disease Activity Index (SSDAI) was ascertained when collected blood [16]. Autoantibody ANA was examined with indirect immunofluorescence; anti-SSA/SSB was tested by enzyme linked immunosorbent assay (ELISA) (Zeus Scientific, Somerville, NJ, USA); and rheumatic factor (RF), IgG, IgA, IgM, C3, C4, CRP were detected by immunoturbidimetry (Roche, Mannheim, Germany). ESR was tested through Westergren’s method.
PBMC preparation and RNA collection
Peripheral blood (8 ml) was obtained from each participant. Total RNA was extracted from peripheral blood mononuclear cells (PBMCs) after lysis with TRIzol reagent. Eligible RNA by quantity and quality testing was used for reverse transcription reaction.
RNA labeling and microarray analysis
Linear RNAs were removed by Rnase R from total RNAs and collected enriched circRNAs, which from individual sample were amplified and transcribed into cRNA using random primer method (Arraystar Inc.). Labeled cRNA was then fragmented and assembled to the circRNA expression microarray slide. Statistically differentially expressed circRNAs between patients and healthy controls were discriminated by Volcano Plot filtering. Similarly, fold change filtering, hierarchical clustering showed different circRNAs expression as well.
CircRNA and miRNA interaction
It is known that circRNAs are able to affect miRNA, which regulates gene expression [17]. In the present study, putative circRNA/microRNA interaction was predicted and miRNA response elements (MREs) for circRNAs were searched for by Arraystar miRNA target prediction software (Arraystar) based on TargetScan [18] and miRanda algorithms [19].
Validation of differentially expressed circRNA by qRT-PCR
To identify the most clinically applicable biomarkers, we selected two circRNAs from the top upregulated and three circRNAs from the top downregulated circRNAs based on the fold change and significance (fold change (FC) > 2.0 and P value < 0.05) from the microarray data for verification. Expression levels of the five circRNAs were confirmed by another 30 paired samples of pSS, SLE patients, and healthy controls by qRT-PCR. qRT-PCR was performed with SYBR Green (QuantiNova™ SYBR Green PCR kit, Qiagen, Shanghai, China). The primer for each gene was summarized in Supplementary Table 1. Relative quantifications of the genes were examined on an Applied Biosystems 7900 Real-Time PCR system as previously described [20].
Statistics
Statistics were performed by SPSS version 10.01 software. Receiver operating characteristic curve (ROC) was used for evaluating diagnostic value of circRNA which was different between pSS and SLE patients, healthy controls. P value (two-tailed) < 0.05 was considered as statistically significant. With respect to selection of differentially expressed circRNAs, the P value < 0.05 and absolute fold change (FC) > 2.0 were considered statistically significant. Sample size was assessed by power and sample size calculation version 3.1.6 software (http://biostat.mc.vanderbilt.edu/PowerSampleSize).
Results
CircRNA expression profiles
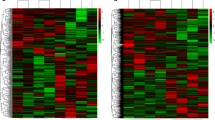
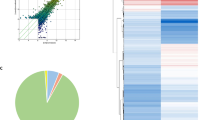
In this case-control study, the sample size was 35 to detect a twofold increased risk assuming an α value of 0.05. CircRNA expression profiles were determined from 5 pSS, SLE, and healthy volunteers. Results showed that circRNA expression profiles in pSS, SLE patients differed from that in control groups, respectively (Fig. 1a). As shown in a box plot (Fig. 1b), distribution of the normalized intensities from all the datasets in the detected subjects were identical. Scatter plot evaluated circRNA expression variation within two groups (Fig. 1c), which was further identified via the volcano plot filtering (Fig. 1d). Overall, there were 234 circRNAs differentially expressed among the pSS patients and controls, 129 were increased and 105 were reduced in pSS patients (data not shown). The top eight elevated and six reduced circRNAs sorted by their FC values are listed in supplementary table 2. Similarly, 148 circRNAs were differentially expressed in SLE patients and controls, 19 were elevated and 129 were reduced in SLE patients (data not shown).
CircRNA expression profile in pSS, SLE patients, and healthy controls. CircRNA expression was determined by microarray analysis in five primary Sjögren’s syndrome (pSS), five systemic lupus erythematosus (SLE) patients, and five healthy controls. a Hierarchical cluster analysis. Red indicates high relative expression and green indicates low relative expression. b Box plot shows the variations in circRNA expression. The distributions were nearly the same after normalization. c Scatterplot figuratively expresses the changes in circRNA expression. CircRNAs in the scatter plot above the top green line and below the bottom green line suggest more than a twofold change of circRNAs between the paired groups (a, pSS patients versus healthy controls; b, SLE patients versus healthy controls). d Volcano plot was prepared according to the fold change (FC) and P value (FC > 2, P < 0.05). The red blocks indicate differentially expressed circRNAs, and gray blocks represent circRNAs with no difference in their expression (a, pSS patients versus healthy controls; b, SLE patients versus healthy controls)
Interaction between differential expression of circRNAs and miRNAs
Utilizing the Arraystar’s homemade miRNA target prediction software based on miRanda, miRNAs paring to circRNAs were predicted and the top five predicted miRNA targets of circRNA were selected. The top eight upregulated and six downregulated circRNAs and their target miRNAs with respect to pSS patients are listed in supplementary table 2. The results included MRE sequences, miRNA seed types, AU-richness near seed sequences, and relative positions of MREs in the linearized sequences of circRNAs. For instance, the potential miRNA targets for hsa_circRNA_001264 include hsa-miR-18a-3p, -607, -632, -654-3p, -10b-3p (Supplementary fig. 1). Correlation between hsa_circRNA_001264 and hsa-miR-18a-3p showed that the 72th–76th nucleotides are beginning from the 5′ terminal region in the higher expressed gene ST6GALNAC3, and the nucleotides were completely and represented a match to the seed region of miR-18a-3p in the binding mode of 8mer.
Validation of the circRNA profile by qRT-PCR
We selected two upregulated (hsa_circRNA_001264, _104121) and three downregulated circRNAs (hsa_circRNA_045355, hsa_circRNA_103461, hsa_circRNA_105034) for further validation in pSS patients. In addition, we validated expression of these circRNAs in 30 lupus patients. The primers are listed in supplementary Table 1. mRNA expression of hsa_circRNA_001264 and hsa_circRNA_104121 was strongly elevated in pSS patients as compared to that in SLE, healthy controls (P < 0.01), whereas mRNA expression of these circRNAs in lupus patients did not strongly differ from controls (P > 0.05) (Fig. 2a and b). Moreover, mRNA expression of hsa_circRNA_045355 was reduced in pSS patients as compared to that in SLE patients, healthy controls (P < 0.01), whereas mRNA expression of hsa_circRNA_103461 and hsa_circRNA_105034 was comparable among pSS, SLE patients, and controls (Fig. 2c–e). Expression of hsa_circRNA_045355, hsa_circRNA_103461, and hsa_circRNA_105034 was not significantly different between SLE patients and healthy controls (P > 0.05) (Fig. 2c–e).
Confirmation of the differential expression of circRNAs by qRT-PCR. Top two upregulated (hsa_circRNA_001264, hsa_circRNA_104121) and three downregulated (hsa_circRNA_045355, hsa_circRNA_103461, hsa_circRNA_105034) circRNAs in pSS patients were selected for validation. a–e Expression levels of five circRNAs were validated by qRT-PCR in the PBMCs from 30 pSS, 30 SLE patients, and 30 healthy controls. The Kruskal-Wallis H test was used
To evaluate the potential of these circRNAs as biomarkers for pSS patients, ROC analysis was conducted. We noted that AUC, sensitivity, and specificity for hsa_circRNA_001264 was 0.860, 80.0%, and 73.3% when pSS patients versus SLE patients and 0.926, 86.7%, and 90.0% when pSS patients versus healthy controls (Fig. 3a and b). With respect to hsa_circRNA_104121, AUC, sensitivity, and specificity were 0.833, 73.3%, and 70.0% when pSS patients versus SLE patients and 0.857, 86.7%, and 70.0% while pSS patients versus healthy controls (Fig. 3c and d). When pSS patients versus SLE patients according to hsa_circRNA_045355, AUC, sensitivity, and specificity were 0.804, 76.7%, and 66.7% and 0.733, 70.0%, and 63.3% for hsa_circRNA_045355 when pSS patients versus healthy controls (Fig. 3e and f). Results of ROC analysis of hsa_circRNA_103461 and hsa_circRNA_105034 were not significant when pSS patients compared with SLE patients and healthy controls (data not shown).
Receiver operating characteristic curve (ROC) analysis of hsa_circRNA_001264, hsa_circRNA_104121, and hsa_circRNA_045355 in PBMCs of pSS patients. a, b Relative mRNA level of hsa_circRNA_001264 in pSS patients versus SLE patients, healthy controls. c, d ROC analysis of hsa_circRNA_104121 in pSS patients when compared to SLE patients and healthy controls. e, f ROC analysis of hsa_circRNA_045355 for pSS patients. AUC values are given on the graphs
Correlation between validated circRNAs and pSS patients’ clinical, laboratory parameters
Early pSS patients (disease duration < 18 months) showed significantly higher expression of hsa_circRNA_001264 as compared to patients with longer disease duration (disease duration > 18 months) (P = 0.008) (Table 2). pSS patients with renal involvement, arthritis also showed higher levels of hsa_circRNA_001264 when compared to that in patients without these clinical parameters (P < 0.001, P = 0.002). Similarly, early pSS patients showed higher expression of hsa_circRNA_104121 compared to that in patients with longer disease duration (P = 0.002). pSS patients with arthritis reported elevated mRNA expression of hsa_circRNA_104121 compared with the patients without the parameter. On the contrary, expression of hsa_circRNA_045355 was lower in early pSS patients compared to that in longer pSS patients (P = 0.020). pSS patients with renal involvement also displayed lower expression of hsa_circRNA_045355 (P = 0.018) (Table 2).
With respect to circRNA expression and laboratory parameters, pSS patients with positive ANA, anti-SSA, and anti-SSB showed higher expression of hsa_circRNA_001264 when compared to that in patients with negative parameters (P = 0.009, P = 0.002, P = 0.026) (Supplementary Table 3). Similarly, patients with positive ANA, anti-SSA, and anti-SSB showed higher expression of hsa_circRNA_104121 when compared to that in patients with negative parameters (P = 0.005, P = 0.008, P = 0.030). By contrast, pSS patients with positive anti-SSA reported lower mRNA expression of hsa_circRNA_045355 compared to that in anti-SSA-negative patients (P = 0.009). Interestingly, there is strong relationship among hsa_circRNA_001264 and hsa_circRNA_104121 mRNA expressions and SSDAI (all P < 0.05) (Table 3). hsa_circRNA_001264 expression also correlated with ESR (rs = − 0.403, P = 0.027). Expression of hsa_circRNA_045355 was negatively related to SSDAI (rs = − 0.624, P < 0.001) (Table 3). The expression of hsa_circRNA_103461 and hsa_circRNA_105034 was not significantly related to pSS patients’ clinical, laboratory parameters (data not shown).
Discussion
pSS is a complex disorder with a focal chronic inflammation of glandular parenchyma, with chronic and persistent involvement of major salivary gland remaining a key element of the disease. Although anti-inflammatory strategies might have effect for correcting the disorder [21,22,23], the molecular mechanisms contributed to inflammation in the disease need clear elucidation.
CircRNAs are accepted to stably exist in cells for a long time [24] and extensively found in mammalian cells, and sometimes, there are over 10 times more circRNAs than linear RNAs [25]. Moreover, circRNAs have highly conserved sequences, a stable existence, and tissue-specific expression. All these suggested that circRNAs may be good biomarkers for diseases and may play a role in the pathogenesis of human diseases. Recent findings displayed that certain circRNAs are involved in rheumatic autoimmune diseases such as arthritis and lupus, and some circRNAs may be used as biomarkers [10, 11, 26]. However, relationship among circRNAs and pSS is unknown.
In this study, 234 circRNAs were differently expressed between pSS patients and healthy controls by usage of RNAseq. We selected the top two upregulated (hsa_circRNA_001264, hsa_circRNA_104121) and three downregulated (hsa_circRNA_045355, hsa_circRNA_103461, hsa_circRNA_105034) differential circRNAs for further validation and found that mRNA expression of hsa_circRNA_001264 and hsa_circRNA_104121 was upregulated in pSS patients, and hsa_circRNA_045355 expression was reduced in pSS patients when compared to those in healthy individuals. Interestingly, pSS patients with early disease showed higher expression of hsa_circRNA_001264, and hsa_circRNA_104121 and downregulated expression of hsa_circRNA_045355 compared to those in patients with longer diseases. These circRNAs were partly related to renal involvement, arthritis, and laboratory parameters, including ANA, anti-SSA, anti-SSB, ESR. Dysregulated expression of hsa_circRNA_001264, hsa_circRNA_104121, and hsa_circRNA_045355 was strongly related to disease activity index. These data suggested that the three circRNAs may correlate with the pathogenesis of pSS. However, the clear roles of circRNAs played in pSS clinical, laboratory parameters need to be elucidated in the future. To discuss the potential of circRNAs as biomarkers for pSS, we evaluated the top two upregulated and three downregulated circRNAs for analysis in a validation set. This showed us that hsa_circRNA_001264, hsa_circRNA_104121, hsa_circRNA_045355 may be good diagnostic biomarkers for pSS patients. Furthermore, we discussed expression profile of circRNAs in PBMCs of SLE patients. Results revealed 148 differentially expressed circRNAs. This was different from the previous studies [12, 26]. Li et al. measured circRNA expression profile in T cells from lupus patients and identified 127 differentially expressed circRNAs, while another study assessed circRNA expression profile from plasma samples in lupus patients and reported 207 differentially expressed circRNAs. The inconsistence may relate to different sample sources and sample sizes.
miRNAs are endogenous non-coding small RNAs. They are essential for regulating gene expression, cell development, differentiation, and function, playing important roles in rheumatic autoimmune diseases, such as pSS [27, 28]. CircRNAs can competitively bind and suppress the corresponding miRNAs. Recent investigations have reported some potential effects of cicrRNAs on miRNAs [29]. However, there is no evidence about role of circRNAs and miRNAs on the impaction of pSS. Hsa_circRNA_001264 is significantly upregulated in pSS patients. miR-18a-3p is one of it MREs. In knee anterior cruciate ligament tissues derived from patients with osteoarthritis, miR-18a-3p expression was reduced [30]. Likewise, miR-203a-3p and miR-143-3p were two MREs of hsa_circRNA_104121, and hsa_circRNA_104121 expression was elevated in pSS patients. Expression of salivary miR-203a-3p was more highly expressed in the cases with bad oral health [31]. Expression of miR-143-3p was higher in synovium tissues of RA than that of controls [32]. Inhibition of miR-143-3p suppressed cell proliferation, reduced levels of inflammatory cytokines, and promoted apoptosis. These data indicated that circRNAs hsa_circRNA_001264 and hsa_circRNA_104121 may bind to downstream miRNAs and therefore play a potential role in the pathogenesis of pSS, such as the clinical parameter arthritis. However, the clear role needs to elucidate in the future.
There are some limitations in this study. First, sample size is relatively small. These data need to be confirmed in large-scale studies and with different races. Second, to determine whether hsa_circRNA_001264, hsa_circRNA_104121, and hsa_circRNA_045355 can be biomarkers for pSS, it should be evaluated for their ability to effectively distinguish pSS from other rheumatic autoimmune diseases, such as rheumatoid arthritis. Third, functional analysis is needed; for instance, we can discuss the performance of circRNAs in regulating B cells that have confirmed significant roles in pSS.
Overall, this study revealed aberrant expression profiles of circRNAs in pSS patients for the first time. The findings may enhance the knowledge regarding the role of circRNAs in pSS. CircRNAs may have value in pSS diagnosis and have potential clinical significance.
Abbreviations
- CircRNA:
-
Circular RNA
- BMI:
-
Body mass index
- RF:
-
Rheumatoid factor
- ANA:
-
Anti-nuclear antibody
References
Shi H, Cao N, Pu Y, Xie L, Zheng L, Yu C (2016) Long non-coding RNA expression profile in minor salivary gland of primary Sjögren’s syndrome. Arthritis Res Ther 18:109. https://doi.org/10.1186/s13075-016-1005-2
Gajewski TF, Schreiber H, Fu YX (2019) Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14:1014–1022. https://doi.org/10.1038/ni.2703
Gierut A, Perlman H, Pope RM (2010) Innate immunity and rheumatoid arthritis. Rheum Dis Clin N Am 36:271–296. https://doi.org/10.1016/j.rdc.2010.03.004
Li Y, Shi X (2013) MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol Immunol 10:65–71. https://doi.org/10.1038/cmi.2012.55
O'Connell RM, Rao DS, Baltimore D (2012) microRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312. https://doi.org/10.1146/annurev-immunol-020711-075013
Bolha L, Ravnik-Glavač M, Glavač D (2017) Circular RNAs: biogenesis, function, and a role as possible cancer biomarkers. Int J Genomics 2017:6218353. https://doi.org/10.1155/2017/6218353
Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23:1743–1748. https://doi.org/10.1101/gad.1812509
Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284:5731–5741. https://doi.org/10.1074/jbc.M804280200
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858. https://doi.org/10.1093/nar/gkw027
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi GP, Yang M (2017) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem 42:651–659. https://doi.org/10.1159/000477883
Zheng F, Yu X, Huang J, Dai Y (2017) Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep 16:8029–8036. https://doi.org/10.3892/mmr.2017.7638
Li LJ, Zhu ZW, Zhao W, Tao SS, Li BZ, Xu SZ, Wang JB, Zhang MY, Wu J, Leng RX, Fan YG, Pan HF, Ye DQ (2018) Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology 155:137–149. https://doi.org/10.1111/imm.12940
Wu Y, Zhang Y, Zhang Y, Wang JJ (2017) CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol Int 41:1283–1289. https://doi.org/10.1002/cbin.10761
Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, Daniels TE, Fox PC, Fox RI, Kassan SS, Pillemer SR, Talal N, Weisman MH (2002) Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American-European consensus group. Ann Rheum Dis 61:554–558. https://doi.org/10.1136/ard.61.6.554
Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25:1271–1277
Vitali C, Palombi G, Baldini C, Benucci M, Bombardieri S, Covelli M, Del Papa N, De Vita S, Epis O, Franceschini F, Gerli R, Govoni M, Bongi SM, Maglione W, Migliaresi S, Montecucco C, Orefice M, Priori R, Tavoni A, Valesini G (2007) Sjögren’s syndrome disease damage index and disease activity index: scoring systems for the assessment of disease damage and disease activity in Sjögren's syndrome, derived from an analysis of a cohort of Italian patients. Arthritis Rheum 56:2223–2231. https://doi.org/10.1002/art.22658
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283. https://doi.org/10.3389/fgene.2013.00283
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS (2003) MicroRNA targets in drosophila. Genome Biol 5:R1 http://genomebiology.com/2003/5/1/R1. Accessed 1 Jan 2019
Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13:271–282. https://doi.org/10.1038/nrg3162
Yuan ZC, Wang JM, Huang AF (2019) Elevated expression of interleukin-37 in patients with rheumatoid arthritis. Int J Rheum Dis 2019:1123–1129. https://doi.org/10.1111/1756-185X.13539
Rodrigues AR, Soares R (2017) Inflammation in Sjögren's syndrome: cause or consequence? Autoimmunity 50:141–150. https://doi.org/10.1080/08916934.2017.1280027
Barone F, Colafrancesco S (2016) Sjögren’s syndrome: from pathogenesis to novel therapeutic targets. Clin Exp Rheumatol 34:58–62
Carsons SE, Vivino FB, Parke A, Carteron N, Sankar V, Brasington R, Brennan MT, Ehlers W, Fox R, Scofield H, Hammitt KM, Birnbaum J, Kassan S, Mandel S (2017) Treatment guidelines for rheumatologic manifestations of Sjögren's syndrome: use of biologic agents, management of fatigue, and inflammatory musculoskeletal pain. Arthritis Care Res (Hoboken) 69:517–527. https://doi.org/10.1002/acr.22968
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148. https://doi.org/10.1016/j.canlet.2015.06.003
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211. https://doi.org/10.1038/nrm.2015.32
Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y (2018) Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta 480:17–25. https://doi.org/10.1016/j.cca.2018.01.026
Chen JQ, Papp G, Szodoray P, Zeher M (2016) The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev 15:1171–1180. https://doi.org/10.1016/j.autrev.2016.09.003
Wang-Renault SF, Boudaoud S, Nocturne G, Roche E, Sigrist N, Daviaud C, Bugge Tinggaard A, Renault V, Deleuze JF, Mariette X, Tost J (2018) Deregulation of microRNA expression in purified T and B lymphocytes from patients with primary Sjögren’s syndrome. Ann Rheum Dis 77:133–140. https://doi.org/10.1136/annrheumdis-2017-211417
Li X, Yang L, Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442. https://doi.org/10.1016/j.molcel.2018.06.034
Li B, Bai L, Shen P, Sun Y, Chen Z, Wen Y (2017) Identification of differentially expressed microRNAs in knee anterior cruciate ligament tissues surgically removed from patients with osteoarthritis. Int J Mol Med 40:1105–1113. https://doi.org/10.3892/ijmm.2017.3086
Kobayashi T, Tomofuji T, Machida T, Yoneda T, Ekuni D, Azuma T, Maruyama T, Hirose A, Morita M (2017) Expression of salivary miR-203a-3p was related with Oral health-related quality of life in healthy volunteers. Int J Mol Sci 18:1263. https://doi.org/10.3390/ijms18061263
Yang Z, Wang J, Pan Z, Zhang Y (2018) miR-143-3p regulates cell proliferation and apoptosis by targeting IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway in rheumatoid arthritis. Exp Ther Med 15:3781–3790. https://doi.org/10.3892/etm.2018.5907
Funding
This study was funded by Sichuan Provincial Science and Technology Program (2019YJ0540), the Key Project of Sichuan Education Department (17ZA0430).
Author information
Authors and Affiliations
Contributions
LS, WX, XL, LF, and AH designed this study. LS, WX, XL, LF, and AH collected data. LS, WX, LF, and AF did laboratory test and statistical analysis. LS, WX, LF, and AF wrote the paper.
Corresponding authors
Ethics declarations
Disclosure
None.
Statement on consent for publication
The whole authors permitted submission of the paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary figure 1
Detailed annotation for circRNA-miRNA interactions. Representative notation example shows the complementary situation of hsa_circRNA_001264. (PNG 1257 kb)
Supplementary table 1
Primer sequence of internal reference and the 5 circRNAs. (DOCX 16 kb)
Supplementary table 2
The top eight elevated, six reduced circRNAs differed in pSS patients. (DOCX 20 kb)
Supplementary table 3
Association of hsa_circRNA_001264, hsa_circRNA_104121, hsa_circRNA_045355 expression and pSS patients’ laboratory characteristics. (DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Su, LC., Xu, WD., Liu, XY. et al. Altered expression of circular RNA in primary Sjögren’s syndrome. Clin Rheumatol 38, 3425–3433 (2019). https://doi.org/10.1007/s10067-019-04728-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-019-04728-6