Abstract
Introduction/objectives
Articular cartilage is the target tissue of osteoarthritis (OA), and because it lacks capillary networks, the microenvironment is hypoxic. Hypoxia inducible factor-1 alpha (HIF-1α) regulates the homeostasis of this tissue. The aim of this study was to investigate whether genetic polymorphisms of the HIF-1α signaling pathway are involved in the development of knee OA.
Method
We performed a case-control association study and genotyped 134 knee OA patients and 267 healthy controls. All participants were genotyped in order to evaluate 42 SNPs from 22 genes involved in the HIF-1α signaling pathway using the OpenArray technology. Gene-gene interactions (epistasis) were analyzed using the multifactor dimensionality reduction (MDR) method.
Results
The MDR analysis showed epistasis between AKT2 (rs8100018) and IGF1 (rs2288377), AKT2 (rs8100018) and IGF1 (rs35767), IGF1 (rs35767) and COL2A1 (rs1793953), and between GSK3B (rs6438552) and IGF1 (rs35767) polymorphisms, with information gain values of 21.24%, 8.37%, 9.93%, and 5.73%, respectively. Additionally, our model allowed us to identify high- and low-risk genotypes among COL2A1 rs1793953, GSK3B rs6438552, AKT2 rs8100018, and IGF1 rs35767 polymorphisms.
Conclusions
Knowing the interactions of these polymorphisms involved in HIF-1α signaling pathway could provide a new diagnostic support tool to identify individuals at high risk of developing knee OA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary osteoarthritis (OA) is a disorder involving movable joints characterized by cartilage degradation, bone remodeling, osteophyte formation, joint inflammation, and loss of normal joint function that can culminate in illness [1]. Worldwide estimates indicate that 9.6% of men and 18% of women ≥ 60 years old suffer from symptomatic OA [2,3,4]. Similar to different ethnic groups, OA is the most common form of human arthritis among Mexicans; its incidence is increasing steadily due to the current demographic, epidemiological, and social transitions along with the pandemic of overweight and obesity in this ethnic group [5].
Articular cartilage is the target tissue of OA, and because it lacks capillary networks, the microenvironment is hypoxic [6]. In physiological conditions, oxygen concentration in articular cartilage varies from 0.5 to 10%. Hypoxia inducible factor-1α (HIF-1α) plays a fundamental role in maintaining the homeostatic conditions of articular cartilage [7,8,9,10,11]. Under normoxia, its specific proline residues 402 and 564 are hydroxylated in the oxygen-dependent degradation domain by prolyl-hydroxylases (PHDs) to form a complex with the von Hippel-Lindau (VHL) factor; in turn, this complex is subsequently degraded in the proteasome [12, 13]. However, under hypoxic conditions, the activity of PHDs decreases, stabilizing HIF-1α, which accumulates in the cytoplasm and is phosphorylated by MAPK [11, 14,15,16]. On the other hand, it has been that the inhibition or depletion of GSK-3 induces HIF-1α, while the overexpression of GSK-3β reduces the expression of HIF-1α [17]. Upon phosphorylation, HIF-1α translocates to the nucleus and binds to specific DNA sequences (5′TAGCGTGH3′) present in promoter regions of genes for their subsequent expression [18, 19]. Among many others, these target genes include NOS2, VEGF, EPO, GLUT1, IGF2, SOX9, and COL2A1. Transcription of such target genes has the potential role of maintaining the chondroprotective functions that are challenged by the detrimental conditions occurring in the OA joint environment [20,21,22,23].
From a genetic standpoint, several studies suggest associations between single-nucleotide polymorphisms (SNPs) and knee OA [24, 25]. Nevertheless, most of them were assessed individually, in contrast to joint assessments through gene-gene interactions (epistasis), which could provide more information regarding their role [26]. The identification and characterization of gene-gene and gene-environment interactions have been limited primarily due to a lack of powerful statistical methods, and particularly because of small sample sizes, which has been a challenge for geneticists. In this sense, the multifactor dimensionality reduction (MDR) method does not require a model as such, given that no genetic models are assumed, neither is it parametric, as no parameters are estimated [27, 28]. The generalized MDR (GMDR) method is an extension from MDR and allows an adjustment for discrete and quantitative covariables and can be applied to both dichotomous and continuous phenotypes in several study designs based on population [29].
Interactions between multiple loci of different genes could be the foundation of the knee OA genetic origin. Therefore, this study is focused on evaluating whether interactions between several genetic variants of HIF-1α signaling pathway are associated with knee OA in the Mexican population.
Materials and methods
Study design and population
Four hundred and one unrelated Mexican-Mestizo individuals were recruited from September 2013 to September 2016 period for this case control-study. One hundred thirty-four of them were primary knee OA patients: 94 from the Instituto Nacional de Rehabilitación “Luis Guillermo Ibarra Ibarra” (INRLGII), and 40 from the Rheumatology Department of the Hospital Civil de Guadalajara “Fray Antonio Alcalde” (Ref. J45703-M CONACYT). The knee OA diagnosis was based on the American College of Rheumatology criteria [30], which included primary OA with any symptoms, and radiographic signs of OA according to the Kellgren-Lawrence (KL) score (≥ 2); the clinical examination and radiographic evaluation were performed by a qualified radiologist-rheumatologist. One hundred and fifty healthy employees from INRLGII and 117 healthy subjects from Guadalajara with no symptoms or signs of knee OA, other types of arthritis, or any painful condition of the joint were recruited as controls. The control subjects were selected among individuals with no personal and family history of OA. Knee radiographs from controls were obtained consecutively to rule out subclinical OA, and those who were grade one or less were considered. Other etiologies causing knee diseases, such as inflammatory arthritis (rheumatoid arthritis -RA- or any other autoimmune disease), post-traumatic or post-septic arthritis, poliomyelitis, and skeletal dysplasia, were excluded. This study meets all criteria contained in the Declaration of Helsinki and was approved by the Ethics and Research Committee of the Instituto Nacional de Rehabilitación (Ref. INR-18/13). All participants signed an informed consent letter; additionally, information on age, gender, weight, body mass index (BMI), and birth place was obtained. All participants were > 40 years and were geographically matched (Mexico City and neighboring states), and to have parents and grandparents born in the same geographical region.
SNPs selection and genotyping
Using a case-control design, we sought to assess the contribution of SNPs involved in the HIF-1α signaling pathway previously reviewed [31]; in addition, we include SNPs that have not been studied in order to know their involvement in OA. A total of 42 SNPs were genotyped in cases and controls and with a population frequency greater than 1% in Mexico population. SNPs selection was supported on information from the http://browser.1000genomes.org/index.html, http://www.ncbi.nlm.nih.gov/projects/SNP/ and http://www.genome.jp/kegg-bin/show_pathway?hsa04066 sources. The selection order of the SNPs was first in promoter regions, followed by exons and introns; also, these SNPs should not be in linkage disequilibrium (LD). Seven SNPs in genes that activate the HIF-1α system, 13 SNPs in genes that interact directly with HIF-1α, and 22 SNPs from genes that are induced by HIF-1α were selected for this study (Table 1). Since the Mexican-Mestizo population is admixed, ancestry informative markers (AIMs) were used to assess whether any association could be confounded due to population stratification (Table 1). A panel of nine AIMs distinguishing mainly Amerindian, African, and European ancestry (δ > 0.44) were genotyped [32, 33].
Genomic DNA was isolated from peripheral blood white cells using a commercial kit based on the salt fractionation method (QIAmp 96 DNA Blood Kit, Qiagen, Hilden, Germany). Genotyping was performed using the OpenArray technology in a QuantStudio 12 K flex System (Thermo Fisher Scientific). Genomic DNA samples were normalized at 50 ng/μl, and 2.5 μl of DNA were mixed with 2.5 μl of TaqMan OpenArray Genotyping Master Mix (Thermo Fisher Scientific) on 384-well plates. Mixes were loaded onto genotyping OpenArray plates previously loaded with the genotyping primers and probes, using the AccuFill System (Thermo Fisher Scientific). Amplification was carried out following the manufacturer’s protocol. Results were analyzed using the TaqMan Genotyper v1.2 software.
Statistical analysis
The clinical variables were evaluated with Student’s t test or Fisher’s exact test, when appropriate, and values were expressed as mean ± SD. Gene and allele frequencies of all polymorphisms were calculated and compared between cases and controls using Fisher’s exacts test. In order to control the global false positive rate, only SNPs with a statistically significant p value on Fisher’s exact test were considered in the multivariate analysis. Associations of each SNP with OA risk were assessed with logistic regression models adjusted by age, gender, BMI, and ancestry, taking into account a co-dominant inheritance model for the SNP. Hardy-Weinberg equilibrium (HWE) was evaluated by calculating the inbreeding coefficient (Fis) using the Genetix v4.05.2 (Université de Montpellier) program with 1000 permutations each loci in both study groups.
The ancestry was analyzed by STRUCTURE software v2.3.4 (Pritchard Lab, Stanford University, USA), to evaluate the effect of population stratification on the associations found of each population k (k = 3) with the genotypes of the nine AIMs mentioned above. This information was included in the logistic regression models to adjust the associations found between the studied polymorphism and OA by individual mix. In addition, we performed a haplotype analysis to determine the joint effect of variants of the same gene on OA development. All the statistical analyses were performed using the statistical package STATA v14.0 (Stata Corp, Texas USA), and considering an α = 0.05 significance level. Finally, in order to study the effect of epistasis, we used the MDR v3.0.2 and GMDR v0.9 statistical packages according the Ritchie’s algorithm [27].
Results
Characteristics of the study population
Demographic and clinical characteristics of knee OA patients and controls are shown in Table 2. In the study groups, cases were significantly older than controls individuals (P < 0.0001, 51.3 ± 13.5 vs 43.6 ± 11.3 years, respectively). Most of the patients were female in both study groups (88.0% in cases and 70.0% in controls, P < 0.0001). The mean BMI of the OA group was significantly higher than the control group (P < 0.0001, 29.2 ± 4.8 vs 26.1 ± 4.8, respectively). There was no difference among patients and controls regarding the place of birth (P = 0.146). The distribution of the studied polymorphisms was consistent with HWE except for HIF1AN rs11292, HIF1A rs11549465, and EGLN1 rs1339894 polymorphisms (Supplementary Table 1).
Association of SNPs of the HIF-1α signaling pathway with OA
After adjusting by age, gender, BMI, and admixture in a logistic regression model, the genotype and allele frequencies of ten SNPs significantly associated are presented in Table 3. Genotypes and alleles with low risk against OA were C/C genotype and C allele of AKT2 rs8100018 (OR = 0.17, 95% CI = 0.05–0.55, P = 0.003, and OR = 0.58, 95% CI = 0.38–0.87, P = 0.009, respectively), C/T genotype and T allele of AGER rs2070600 (OR = 0.05, 95% CI = 0.00–0.47, P = 0.008, and OR = 0.23, 95% CI = 0.08–0.64, P = 0.005, respectively), A/G genotype of HIF1AN rs11292 (OR = 0.37, 95% CI = 0.14–0.96, p = 0.04), A/A genotype and A allele of EGLN1 rs1339894 (OR = 0.05, 95% CI = 0.00–0.45, P = 0.007, and OR = 0.39, 95% CI = 0.22–0.70, P = 0.001, respectively), A/A genotype of VEGFA rs1570360 (OR = 0.31, 95% CI = 0.10–0.93, p = 0.03), and G/A genotype of COL2A1 rs1793953 (OR = 0.48, 95% CI = 0.28–0.82, P = 0.008). On the other hand, genotypes and alleles with high risk to development OA wer: A/G genotype of GSK3B rs6438552 (OR = 2.58, 95% CI = 1.16–4.45, P = 0.01), C/T genotype and T allele of HIF1A rs11549465 (OR = 3.14, 95% CI = 1.82–5.42, P = 0.000, and OR = 2.07, 95% CI = 1.33–3.23, P = 0.001, respectively), A/T genotype and T allele of IGF1 rs2288377 (OR = 1.86, 95% CI = 1.08–3.20, P = 0.02, and OR = 1.63, 95% CI = 1.01–2.63, P = 0.04, respectively), and G/A genotype and A allele of IGF1 rs35767 (OR = 2.00, 95% CI = 1.17–3.42, P = 0.01, and OR = 1.51, 95% CI = 1.02–2.25, P = 0.03, respectively).
Evaluation of gene-gene interactions: MDR
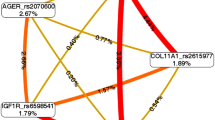
Table 4 summarizes the results of exhaustive MDR analysis, which analyzes all possible combinations of the studied polymorphisms. According to the MDR analysis, the best models include the AKT2 (rs8100018) and IGF1 (rs2288377) polymorphisms. This model had a balanced accuracy test of 0.7678, a consistency of cross-validation of 10/10, and an interaction P value = 0.0010. Figure 1 shows the interaction map of the studied polymorphisms, based on entropy measures among individual variables. A strong interaction effect was observed between AKT2 (rs8100018) and IGF1 (rs2288377), AKT2 (rs8100018) and IGF1 (rs35767), IGF1 (rs35767) and COL2A1 (rs1793953), and between GSK3B (rs6438552) and IGF1 (rs35767) polymorphisms with information gain values of 21.24%, 8.37%, 9.93%, and 5.73%, respectively. The gene-gene interaction of the ten associated polymorphisms is shown in the interaction dendogram (Supplementary Fig.1). Moreover, our model allowed us to identify interactions in high-risk genotypes of the COL2A1 (rs1793953), GSK3B (rs6438552), and IGF1 (rs35767) polymorphisms, and the most representative were (GA + AG + GA), (GA + GG + GA), and (GG + AG + GG), respectively; and low-risk genotypes [(GA + AA+GA), (GA + AG + GG), and (GA + GG + GG)], respectively. Likewise, we identify interactions in high-risk genotypes of the AKT2 (rs8100018) and IGF1 (rs2288377) polymorphisms [(GG + AA) and (GC + AT)], respectively; and low-risk genotypes [(GC + AA) and (GG + AT)], respectively (Fig. 2).
Interaction map for knee OA risk. The interaction model describes the percentage of the entropy (information gain) that is explained by each factor or two-way interaction. Values inside nodes indicate information gain of individual attributes or main effects, whereas values between nodes show information gain of pairwise combinations of attributes or interaction effects. Positive entropy (plotted in red or orange) indicates interaction, which can be interpreted as a synergistic or nonadditive relationship; while negative entropy (plotted in yellow-green) indicates independence or additivity (redundancy)
Distribution of high-risk and low-risk genotypes in the best two- and three-locus model. The distribution shows high-risk (dark shading) and low-risk (light shading) genotypes associated with knee OA in the two- and three-locus interaction detected by MDR analysis. The percentage of osteoarthritic subjects (left black bar in boxes) and control subjects (right hatched bar in boxes) is shown for each two- and three-locus genotype combination. Boxes were labeled as high-risk if the ratio of the percentage of cases to controls met or exceeded the threshold of 1.0. Boxes were labeled as low-risk if the threshold was not exceeded. Based on the pattern of high-risk and low-risk genotypes, this two- and three-locus model is evidence of gene-gene interaction
Haplotype analysis
In regard to the haplotype analysis, we observed that the CTG (rs2057482, rs11549465, and rs11549467, respectively) and AT (rs35767 and rs228377, respectively) haplotypes of the HIF1A and IGF1 genes, respectively, were found to be associated with an increased risk of developing (OR = 2.59, P = 0.004, 95% CI = 1.36–4.94 and OR = 1.69, P = 0.038, 95% CI = 1.02–2.80, respectively) (Supplementary Table 2).
Discussion
OA is the most common joint disease, imposing a major economic burden to health systems due to the costs associated with healthcare and disability [34]. Several studies have been performed aimed to identify potential genes of therapeutic targets [35]. It is well-known that knee OA pathogenesis is multifactorial, and its complexity is primarily due to its polygenic nature. Given this polygenic nature, it has been difficult to prove gene-gene interactions associated with knee OA; in this sense, MDR has been applied to identify gene-gene interactions conferring susceptibility to common multifactorial diseases, including hypertension, bladder cancer, type 2 diabetes, and RA [36]. To date, only two published reports have evaluated gene-gene interactions by the MDR method in knee OA, which allow the identification of predictive models for the disease development based on the analyzed pathways (TGF-β/Smad3 and ADIPOQ/PON1) [37, 38]. In the present study, we applied the MDR method to assess the epistasis of genes related to the HIF-1α signaling pathway due to its central participation in the articular cartilage homeostasis.
Our main findings reveal important gene-gene interactions between the AKT2, IGF1, COL2A1, and GSK3B genes and knee OA. HIF-1α expression is regulated through the PI3K/Akt pathway, and both kinases are important in cell survival and apoptosis; especially, it has been shown that apoptosis of chondrocytes can be regulated by this signaling pathway, which is closely related to the occurrence and development of osteoarthritis [39, 40]. In our study, we observed that the carriers of the G/G homozygous genotype and the G minor allele of the AKT2 rs8100018 polymorphism showed a significant association with a lower risk to knee OA development. To our knowledge, data on the associations between common genetic variations in AKT2 gene and knee OA are scarce. But in pathologies such as rectal cancer, it has been observed that the rs8100018 variant is associated with low risk in progress to cancer, suggesting that this variant might play an important role in the AKT2 function [41].
On the other hand, the insulin-like growth factor-1 (IGF-1) is a small 70-amino acid polypeptide mediator with a potent anabolic impact on cartilage homeostasis. IGF-1 is expressed in cartilage, where it can act in a paracrine and autocrine manner to stimulate cartilage extracellular matrix (ECM) synthesis as well as inhibit matrix degradation [42, 43], and it has a close relationship in the expression of HIF-1α under hypoxic conditions such as occurrence in articular cartilage [44]. In our study, we evaluated the rs35767 and rs2288377 polymorphisms of the IGF1 gene, and we observed that the carriers of the heterozygous genotype and the minor allele in both polymorphisms have higher risk to develop OA. Today, the role of these polymorphisms in the development of OA is not clear. In other pathologies such as osteoporosis, the rs35767 polymorphism has also been associated with risk, especially with low levels of bone mineral density of the femoral neck [45]; however, in the study performed by Chen YC et al., they found that the rs2288377 polymorphism was not associated with osteoporosis risk [46]. In view of these reports, our results may help to elucidate the role that plays the rs35767 and rs2288377 polymorphisms in pathologies that affect the joint and adjacent tissues, but more studies are needed to support it.
Also, we observed that the rs1793953 polymorphism of the COL2A1 gene was associated with protection against OA. It is known that this gene codifies for the alpha chain of type II collagen, which is the main component of the ECM of the articular cartilage. Alterations in this gene have been associated with OA and early onset family OA, among other cartilage disorders [47]. In the study performed by Gálvez-Rosas et al., they analyzed a polymorphic site in the COL2A1 gene of primary knee OA patients and observed a significant association with KL grade 4 patients [48]. Moreover, Valdes et al. analyzed the rs1635560 polymorphism of the COL2A1 gene in OA patients and found an association with a decrease in knee OA risk, but only among male patients (OR = 0.68, P < 0.005) [49]. Deng Y et al. analyzed the rs1793953 polymorphism of the COL2A1 gene in intervertebral disc degeneration patients, and they found that the carriers of the A/A homozygous genotype and of the A minor allele showed a significant association with a lower risk of developing this disease (P = 0.004 and P = 0.010, respectively) [50]. The controversy of these results is highly interesting, suggesting for instance a dual role of the gene in the disease, or even a possible interaction with environmental or genetics factors not taken into account in the latter studies. Thus, it is necessary to explore other polymorphic variants in COL2A1 in our population and elucidate their involvement in OA.
Finally, in the present work, we evaluated the rs6438552 polymorphism of the glycogen synthase kinase-3B (GSK3B) gene in knee OA patients, and we observed that the carriers of the heterozygous A/G genotype increase the risk of OA. Several studies have suggested a proinflammatory role for GSK-3 activity based on cytokine profiles during GSK-3 inhibition. GSK-3 inhibition has been demonstrated to ameliorate collagen-induced arthritis and collagen antibody-induced arthritis in mice, which is consistent with a proinflammatory role; however, its activity may have procatabolic or chondroprotective effects depending on the pathologic scenario, with important implications for the proposed use of GSK-3 inhibitors as therapeutic agents in arthritis [51].
The gene-gene interaction analysis allows us to know whether two or more polymorphisms impact OA genetic susceptibility. Our study allowed us to identify gene-gene interactions implemented by MDR with high-degree synergy between AKT2 and IGF1 genes (Fig. 1). Examination of these genes in the interaction model reveals a testable hypothesis for further studies; not only does the evaluation of interactions between genes increase the detection capacity, but it also helps to understand the genetics behind the underlying biological and biochemical pathways of the disease. Another important aspect is that with the MDR method, high-risk and low-risk genotypes were identifying in knee OA patients, suggesting an essential role of the polymorphisms involved in HIF-1α signaling pathway (Fig. 2). Because the MDR method allows the identification of risk predictive models in OA, it can also be used to provide support in preclinical diagnosis; in addition, knowing the mechanisms of interaction, it could help to designed specific therapeutic strategies where several molecular targets should be taken into account for OA.
Finally, the haplotypes analysis makes it possible to evaluate whether there are polymorphism blocks (groups) of a single gene that are jointly segregated and might be linked to the disease development. Our results show that the presence of CTG and AT haplotypes of the HIF1A and IGF1 genes are significantly associated (P < 0.05) with knee OA (Supplementary Table 2). The data obtained points out the potential role that these genes play in knee OA development.
It is worth mentioning some strengths of our study. a) The population stratification was not biased, given that we included the ethnicity of each participant in the regression models assessed by AIMs; b) our study is the first that evaluated the wide number of genes related to the HIF-1α signaling pathway among Mexican patients with knee OA; and c) unlike genetic classical analysis, our main approach highlights the importance to evaluate in an integral manner the effect of genetic variants in knee OA.
Yet, it is important to highlight some aspects. We are aware of the limitations of our study; first, our sample size is limited; however, we believe that after performing a multivariate analysis and a rigorous selection of our patients and controls, the presented data reinforce the biological plausibility of the SNPs in the OA. Second, our association study was limited to two populations, so more studies in different populations are needed to support our findings, as well as to evaluate the functionality of the associated SNPs and be able to show evidence of whether they have a causal effect or not. Finally, there are more variants of the same gene that were not analyzed, as well as other genes of the HIF-1α signaling pathway that were not considered and whose impact on OA development is unknown.
Conclusions
We analyzed polymorphisms related to the HIF-1α signaling pathway in Mexican knee OA patients. Knowing the gene-gene interactions of these polymorphisms involved in HIF-1α signaling pathway could provide a new diagnostic support tool to identify individuals at high risk of developing knee OA which can serve as a therapeutic target; additionally, a large-scale study to assess HIF-1α signaling pathway polymorphisms and mechanisms of interaction is needed to clarify the role of HIF-1α polymorphisms in the pathogenesis of knee OA.
References
Kraus VB, Blanco FJ, Englund M, Karsdal MA, Lohmander LS (2015) Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthr Cartil 23(8):1233–1241
Arden N, Nevitt M (2006) Osteoarthritis: epidemiology. Best Pract Res Clin Rheumatol 20(1):3–25
De Filippis L, Gulli S, Caliri A, Romano C, Munaó F, Trimarchi G et al (2004) Epidemiology and risk factors in osteoarthritis: literature review data from "OASIS" study. Reumatismo 56(3):169–164
Woolf AD, Pfleger B (2003) Burden of major musculoskeletal conditions. Bull World Health Organ 81(9):646–656
Peláez-Ballestas I, Sanin LH, Moreno-Montoya J, Alvarez-Nemegyei J, Burgos-Vargas R, Garza-Elizondo M et al (2011) Epidemiology of the rheumatic diseases in Mexico. A study of 5 regions based on the COPCORD methodology. J Rheum Suppl 86:3–8
Sophia Fox AJ, Bedi A, Rodeo SA (2009) The basic science of articular cartilage: structure, composition, and function. Sports Health 1(6):461–468
Pfander D, Swoboda B, Cramer T (2006) The role of HIF-1alpha in maintaining cartilage homeostasis and during the pathogenesis of osteoarthritis. Arthritis Res Ther 8(1):104
Ströbel S, Loparic M, Wendt D, Schenk AD, Candrian C, Lindberg RL et al (2010) Anabolic and catabolic responses of human articular chondrocytes to varying oxygen percentages. Arthritis Res Ther 12(2):R34
Fernández-Torres J, Hernández-Díaz C, Espinosa-Morales R, Camacho-Galindo J, Galindo-Sevilla NC, López-Macay A et al (2015) Polymorphic variation of hypoxia inducible factor-1 A (HIF1A) gene might contribute to the development of knee osteoarthritis: a pilot study. BMC Musculoskelet Disord 16:218
López-Reyes A, Rodríguez-Pérez JM, Fernández-Torres J, Martínez-Rodríguez N, Pérez-Hernández N, Fuentes-Gómez AJ, Aguilar-González CA, Álvarez-León E, Posadas-Romero C, Villarreal-Molina T, Pineda C, Vargas-Alarcón G (2014) The HIF1A rs2057482 polymorphism is associated with risk of developing premature coronary artery disease and with some metabolic and cardiovascular risk factors. The Genetics of Atherosclerotic Disease (GEA) Mexican Study. Exp Mol Pathol 96(3):405–410
Zhou J, Hara K, Inoue M, Hamada S, Yasuda H, Moriyama H, Endo H, Hirota K, Yonezawa K, Nagata M, Yokono K (2008) Regulation of hypoxia-inducible factor 1 by glucose availability under hypoxic conditions. Kobe J Med Sci 53(6):283–296
Fan L, Li J, Yu Z, Dang X, Wang K (2014) The hypoxia-inducible factor pathway, prolyl-hydroxylase domain protein inhibitors, and their roles in bone repair and regeneration. Biomed Res Int 2014:239356
Sartori-Cintra AR, Mara CS, Argolo DL, Coimbra IB (2012) Regulation of hypoxia-inducible factor-1α (HIF-1α) expression by interleukin-1β (IL-1 β), insulin-like growth factors I (IGF-I) and II (IGF-II) in human osteoarthritic chondrocytes. Clinics (Säo Paulo, Brazil) 67(1):35–40
Maxwell PH (2005) Hypoxia-inducible factor as a physiological regulator. Exp Physiol 90(6):791–797
Görlach A (2009) Regulation of HIF-1 alpha at the transcriptional level. Curr Pharm Des 15(33):3844–3852
Semenza GL (2002) Signal transduction to hypoxia-inducible factor. Biochem Pharmacol 64(5–6):993–998
Flügel D, Görlach A, Michiels C, Kietzmann T (2007) Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol Cell Biol 27(9):3253–3265
Chen N, Chen LP, Zhang J, Chen C, Wei XL, Gul Y, Wang WM, Wang HL (2012) Molecular characterization and expression analysis of three hypoxia-inducible factor alpha subunits, HIF-1α/2α/3α of the hypoxia-sensitive freshwater species, Chinese sucker. Gene 498(1):81–90
Patel SA, Simon MC (2008) Biology of hypoxia-inducible factor-2alpha in development and disease. Cell Death Differ 15(4):628–624
Semenza GL (2000) HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol 88(4):1474–1480
Pfander D, Cramer T, Swoboda B (2005) Hypoxia and HIF-1 alpha in osteoarthritis. Int Orthop 29(1):6–9
Murphy CL, Thoms BL, Vaghjiani RJ, Lafont JE (2009) Hypoxia. HIF-mediated articular chondrocyte function: prospects for cartilage repair. Arthritis Res Ther 11(1):213
Zhang FJ, Luo W, Lei GH (2015) Role of HIF-1α and HIF-2α in osteoarthritis. Joint Bone Spine 82(3):144–147
Fernández-Moreno M, Rego I, Carreira-García V, Blanco FJ (2008) Genetics in osteoarthritis. Current Genomics 9(8):542–547
van Meurs JB, Uitterlinden AG (2012) Osteoarthritis year 2012 in review: genetics and genomics. Osteoarthr Cartil 20(12):1470–1476
Mechanic LE, Luke BT, Goodman JE, Chanock SJ, Harris CC (2008) Polymorphism interaction analysis (PIA): a method for investigating complex gene-gene interactions. BMC Bioinformatics 9:146
Ritchie MD, Hahn LW, Moore JH (2003) Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol 24(2):150–157
Hahn LW, Ritchie MD, Moore JH (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19(3):376–372
Chen GB, Xu Y, Xu HM, Li MD, Zhu J, Lou XY (2011) Practical and theoretical considerations in study design for detecting gene-gene interactions using MDR and GMDR approaches. PLoS One 6(2):e16981
Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M (1986) Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum 29(8):1039–1049
Fernández-Torres J, Martínez-Nava GA, Gutiérrez-Ruíz MA, Gómez-Quiróz LE, Gutiérrez M (2017) Role of hypoxia inducible factor-1α signaling pathway in osteoarthritis: a systematic review. Rev Bras Reumatol Engl Ed 57(2):162–173. https://doi.org/10.1016/j.rbre.2016.07.008
Choundry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL et al (2006) Population stratification confounds genetic association studies among Latinos. Hum Genet 118(5):652–664
Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD (2004) Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet 68(Pt 2):139–152
Contreras-Hernández I, Mould-Quevedo JF, Torres-González R, Goycochea-Robles MV, Pacheco-Domínguez RL, Sánchez-García S et al (2008) Cost-effectiveness analysis for joint pain treatment in patients with osteoarthritis treated at the Instituto Mexicano del Seguro Social (IMSS): comparison of nonsteroideal anti-inflammatory drugs (NSAIDs) vs. cyclooxygenase-2 selective inhibitors. Cost Eff Resour Alloc 6:21
Chapman K, Valdes AM (2012) Genetic factors in OA pathogenesis. Bone 51(2):258–254
Dai H, Charnigo RJ, Becker ML, Leeder JS, Motsinger-Reif AA (2013) Risk score modeling of multiple gene to gene interactions using aggregated-multifactor dimensionality reduction. BioData Min 6(1):1
Su SL, Yang HY, Lee HS, Huang GS, Lee CH, Liu WS, Wang CC, Peng YJ, Lai CH, Chen CY, Lin C, Pan YT, Salter DM, Chen HC (2015) Gene-gene interactions between TGF-β/Smad3 signaling pathway polymorphisms affect susceptibility to knee osteoarthritis. BMJ Open 5(6):e007931
Fernández-Torres J, Martínez-Nava GA, Zamudio-Cuevas Y, Martínez-Flores K, Espinosa-Morales R (2019) Epistasis between ADIPOQ rs1501299 and PON1 rs662 polymorphisms is potentially associated with the development of knee osteoarthritis. Mol Biol Rep 46(2):2049–2058
Semenza GL (2008) Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiol (Bethesda) 24:97–106
Du XA, Wang HM, Dai XX, Kou Y, Wu RP, Chen Q et al (2015) Role of selenoprotein S (SEPS1) -105G>A polymorphisms and PI3K/Akt signaling pathway in Kashin-Beck disease. Osteoarthr Cartil 23(2):210–216
Peng J, Ma W, Zhou Z, Gu Y, Lu Z, Zhang R, Pan Z (2018) Genetic variations in the PI3K/PTEN/AKT/mTOR pathway predict tumor response and disease-free survival in locally advanced rectal cancer patients receiving preoperative chemoradiotherapy and radical surgery. J Cancer 9(6):1067–1077
Urano T, Narusawa K, Shiraki M, Usui T, Sasaki N, Hosoi T et al (2008) Association of a single nucleotide polymorphism in the insulin-like growth factor-1 receptor gene with spinal disc degeneration in postmenopausal Japanese women. Spine (Phila Pa 1976) 33(11):1256–1261
Claessen KM, Ramautar SR, Pereira AM, Smit JW, Biermasz NR, Kloppenburg M (2012) Relationship between insulin-like growth factor-1 and radiographic disease in patients with primary osteoarthritis: a systematic review. Osteoarthr Cartil 20(2):79–76
Xie Q, Xie J, Zhong J, Cun X, Lin S, Lin Y, Cai X (2016) Hypoxia enhances angiogenesis in an adipose-derived stromal cell/endothelial cell co-culture 3D gel model. Cell Prolif 49(2):236–235
Wei YK, Ma HL, Guo YZ, Yang BH, Pang WX (2015) Association of the IGF-1 rs35767 and rs972936 polymorphisms with the risk of osteoporosis in a Chinese postmenopausal female population. Genet Mol Res 14(4):14325–14330
Chen YC, Zhang L, Li EN, Ding LX, Zhang GA, Hou Y, Yuan W (2017) Association of the insulin-like growth factor-1 single nucleotide polymorphisms rs35767, rs2288377, and rs5742612 with osteoporosis risk: a meta-analysis. Medicine (Baltimore) 96(51):e9231
Snelgrove TA, Peddle LJ, Stone C, Nofball F, Peddle D, Squire D et al (2005) Association of COL1A2, COL2A1 and COL9A1 and primary osteoarthritis in a founder population. Clin Genet 67(4):359–360
Gálvez-Rosas A, González-Huerta C, Borgonio-Cuadra VM, Duarte-Salazár C, Lara-Alvarado L, Soria-Bastida d l A et al (2010) A COL2A1 gene polymorphisms is related with advanced stages of osteoarthritis of the knee in Mexican Mestizo population. Rheumatol Int 30(8):1035–1039
Valdes AM, Loughlin J, Oene MV, Chapman K, Surdulescu GL, Doherty M, Spector TD (2007) Sex and ethnic differences in the association of ASPN, CALM1, COL2A1, COMP, and FRZB with genetic susceptibility to osteoarthritis of the knee. Arthritis Rheum 56(1):137–146
Deng Y, Tan XT, Wu Q, Wang X (2017) Correlations between COL2A and aggrecan genetic polymorphisms and the risk and clinicopathological features of intervertebral disc degeneration in a Chinese Han population: a case-control study. Genet Test Mol Biomarkers 21(2):108–115
Litherland GJ, Hui W, Elias MS, Wilkinson DJ, Watson S, Huesa C, Young DA, Rowan AD (2014) Glycogen synthase kinase 3 inhibition stimulates human cartilage destruction and exacerbates murine osteoarthritis. Arthritis Rheumatol 66(8):2175–2187
Acknowledgements
We thank the support provided by Dr. Gustavo Reyes Terán in facilitating the laboratory where we genotyped the samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Written informed consent was obtained from all participants according to the Declaration of Helsinki and the study protocol was approved by ethics committee of the National Research Centre.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Fernández-Torres, J., Martínez-Nava, G.A., Zamudio-Cuevas, Y. et al. Impact of the gene-gene interactions related to the HIF-1α signaling pathway with the knee osteoarthritis development. Clin Rheumatol 38, 2897–2907 (2019). https://doi.org/10.1007/s10067-019-04635-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-019-04635-w