Abstract
Canine generalized progressive retinal atrophy (gPRA) is characterized by continuous degeneration of photoreceptor cells leading to night blindness and progressive vision loss. Until now, mutations in 11 genes have been described that account for gPRA in dogs, mostly following an autosomal recessive inheritance mode. Here, we describe a gPRA locus comprising the newly identified gene coiled-coil domain containing 66 (CCDC66) on canine chromosome 20, as identified via linkage analysis in the Schapendoes breed. Mutation screening of the CCDC66 gene revealed a 1-bp insertion in exon 6 leading to a stop codon as the underlying cause of disease. The insertion is present in all affected dogs in the homozygous state as well as in all obligatory mutation carriers in the heterozygous state. The CCDC66 gene is evolutionarily conserved in different vertebrate species and exhibits a complex pattern of differential RNA splicing resulting in various isoforms in the retina. Immunohistochemically, CCDC66 protein is detected mainly in the inner segments of photoreceptors in mouse, dog, and man. The affected Schapendoes retina lacks CCDC66 protein. Thus this natural canine model for gPRA yields superior potential to understand functional implications of this newly identified protein including its physiology, and it opens new perspectives for analyzing different aspects of the general pathophysiology of gPRA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Generalized progressive retinal atrophy (gPRA) in domestic dogs is characterized by progressive degeneration of the retina leading to night blindness, followed by loss of peripheral and central visual fields analogous to retinitis pigmentosa (RP), the most prevalent group of inherited retinopathies in man. In most canine breeds, the disease is inherited in an autosomal recessive (ar) manner [1]. Until now, causative mutations have been identified in several breeds of dogs with ar-transmitted gPRA [2–8]. A large number of additional canine breeds suffer from gPRA forms for which DNA tests are not yet available [1].
Previously, a gPRA causing locus in Schapendoes dogs was assigned to canine chromosome 20 (CFA20) between markers FH3358 and TL336MS (distance 5.6 Mb), based on a genome-wide scan of five gPRA-informative pedigrees [9]. Initial mutation screening in three candidate genes close to a microsatellite marker (REN93E07; LOD score 4.78) had revealed no causative sequence deviations (op. cit.). Here, we refined the analysis of this 5.6 Mb region with additional microsatellites and reduced the candidate gene region to 4.2 Mb.
The counterpart of this canine genome region maps to human chromosome 3 (HSA 3p21.1). To date, genomic investigations in RP families have not provided evidence for linkage of an RP form to this chromosomal region (A. Gal, personal communication). Altogether, the respective canine region contains 10 additional candidate genes, eight of which are expressed in the human retina to varying degrees. Hence, respective candidate genes had to be screened sequentially in gPRA-affected Schapendoes dogs in order to identify the disease-causing mutation. In order to evaluate the consequences of any sequence deviation in a newly identified gene, the structure and its RNA expression are to be characterized due to limited sequence information available in respective databases for many a region in the canine genome. Furthermore, gene expression is investigated as a prerequisite for evaluating the consequences of the mutation; however, the respective protein had not been investigated to date. Comparing physiological protein expression patterns in dog, man, and mouse may yield evidence for similar localization in retinal cells. Altogether, investigating the cause of gPRA in Schapendoes necessitated an entry to new genomic and protein territories. Hence, this canine gPRA model may yield also entirely new vistas on certain RP forms in man.
Materials and methods
Animals and humans
Blood samples were collected from the general population of purebred Schapendoes. The primary study sample comprised of 112 DNA samples including 19 DNAs of gPRA-affected dogs [9]. In addition, 870 blood samples were supplied by the owners for indirect diagnostic testing for gPRA. Furthermore, blood from 50 gPRA-affected dogs of 29 different breeds [10] was included in these studies with the permission of the owners and in cooperation with the respective breeding organizations. Genomic DNA was extracted from peripheral blood according to standard protocols [11].
For the isolation of retinal RNA and immunohistochemical staining, we obtained an eye of a gPRA-affected, 6-year-old Schapendoes that had shown loss of night vision previously and had severely impaired day-time vision. As the changes in this affected eye could be secondary to the disease process, immunohistochemical and RNA analyses were complemented in normal eyes. Control samples, i.e. retinae of gPRA-free Saarloos Wolfhounds and a healthy Weimaraner were obtained from veterinarians performing euthanasia because of terminal disease. Eyes from euthanized dogs were provided by ophthalmologically experienced veterinarians with informed consent of all owners. The fundus status of the dogs’ eyes was documented prior to euthanasia.
Mice of the C57BL/6 J strain were obtained from Jackson Laboratory (Bar Harbor, ME, USA), bred at the Ruhr-University, Bochum (RUB), and maintained on an artificial 12 h light/dark cycle. For the isolation of DNA, RNA, and protein, mice were anesthetized by asphyxiation in CO2 followed by decapitation. After dissection, tissue samples were snap-frozen in liquid nitrogen and stored at –80°C until used.
All research and animal care procedures were performed according to international guidelines for the use of laboratory animals.
Archived eyes from humans post mortem were obtained in the Department of Anatomy of the RUB following ethical guidelines of the local Ethics Commission of the RUB (no. 3386-09).
Fine mapping and candidate gene screening
PCR primers for mutation analysis (supplementary Table 1) and tailed primers for further genotyping (supplementary Table 1) were designed using the Primer Express software (Applied Biosystems). Genomic DNA was amplified and genotyped on a MegaBACE 1000 (Amersham Biosciences, Freiburg, Germany) according to the manufacturer’s protocol [10]. Exon/intron boundaries of the following genes were reassessed by comparison of human cDNA counterparts with the publicly available dog genome sequences in the UCSC Genome Browser (assembly: dog May 2005): ARF4 (ADP-ribosylation factor 4; NM_001660.3); ASB14 (ankyrin repeat and SOCS box-containing protein 14; NM_001142733); HESX1 (homeobox gene expressed in ES cells; NM_003865); ARHGEF3 (rho guanine nucleotide exchange factor 3, NM_019555); C3ORF63 (human chromosome 3 open reading frame 63; NM_001112736); CCDC66 (coiled-coil domain containing (protein) 66; NM_001012506); and ERC2 (ELKS/RAB6-interacting/CAST family member 2; NM_015576).
Single-strand conformation polymorphism (SSCP) mutation screening [12] was performed for all coding exons and flanking intronic regions of the abovementioned genes in six gPRA affected, four carrier, and six healthy Schapendoes as well as in four Entlebucher Mountain Dogs (two healthy and two afflicted with retinal degeneration [prcd mutation]) and four Kuvasz (two healthy and two afflicted with retinal degeneration [prcd]). Exons with conspicuous banding patterns in electrophoresis were directly sequenced (MegaBACE 1000, Amersham).
In the region surrounding the newly identified CCDC66 mutation in this study (36.3–38.8 Mb), haplotype analyses were performed with microsatellite markers (FK36.9_20, FK37.3_20, REN149D232, REN316E23, MSS335, FK38.6, and FK38.8_20) in order to reveal key cross-over events in the Schapendoes pedigrees. These marker systems are either contained in the minimal screening set 2 [9] or newly developed for fine mapping of CFA20 (supplementary Table 1). The latter microsatellite systems were based on the dog genome sequence (UCSC; May 2005) as identified using the TANDEM Repeats Browser.
DHPLC analyses
The mutation-containing exon 6 of the CCDC66 gene was investigated by denaturing high-performance liquid chromatography (dHPLC; [13, 14]) on a WAVE® system (Cheshire, UK) with the analysis software Navigator 2.2 in all Schapendoes in order to verify the results of the preceding indirect diagnostic test. PCR amplicons from 870 dogs and from an unaffected control (verified by direct sequencing) were mixed in a 1:1 ratio in order to allow formation of heteroduplexes. These mixes were heated to 95°C for 5 min and slowly cooled to 25°C for 45 min in a thermocycler. PCR amplicons were analyzed at a melting temperature of 54.5°C. The resulting chromatograms were subsequently compared to the chromatogram of the unaffected control sample and another control sample containing a polymorphism.
CCDC66 genomic and initial cDNA analyses
Genomic regions of the CCDC66 gene were amplified by long range PCR (expand high fidelity PCR system; Roche, Mannheim). Newly identified sequences of CCDC66 exons 6 and 7 were verified by overlapping cDNA analysis. The specific cDNA used for this analysis was amplified from total retinal RNA with a primer hybridizing in the 3′ UTR of the CCDC66 gene (supplementary Table 1). The PCR products were purified from agarose gels (Illustra GFX PCR DNA and gel band purification kit) and directly analyzed on a capillary DNA sequencer MegaBACE 1000 (Amersham Biosciences, Freiburg, Germany). CCDC66 cDNA sequence analyses as well as nucleotide and protein database searches were performed using the National Center for Biotechnology Databases (Blast Search: http://blast.ncbi.nlm.nih.gov/Blast.cgi; analysis of splice sites with AceView: http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/; http://www.h-invitational.jp/hinv/ahg-db/index.jsp). Protein and cDNA sequences were aligned using the clustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Quantitative RT-PCR
Total RNA from C57BL/6 J mouse tissues (brain, heart, liver, muscle, and retina), from the retinae of a gPRA-affected Schapendoes, an unaffected Saarloos Wolfhound, and a healthy Weimaraner was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Isolation of total RNA from blood of a healthy dog was performed using the Invisorb Blood RNA Mini kit (Invitek, Berlin, Germany). For quantitative real-time PCR analysis, the QuantiTect SYBR Green assay (Qiagen, Hilden, Germany) was used as described by the manufacturer with the iCycler iQ real-time PCR detection system (BioRad, München, Germany). PCR primers were designed using the Primer Express software (Applied Biosystems). In order to avoid increased risk of potentially contaminating genomic DNA, the primers spanned intron/exon borders. CCDC66 cDNA as well as the rod-specific gene GNAT-1, HPRT, and the external standard GAPDH cDNAs were amplified using primers, as detailed in supplementary Table 1. One-step PCR cycling was carried out by reverse transcription at 50°C for 30 min; initial activation step at 95°C for 15 min × 1 cycle; four-step cycling at 94°C for 15 s; at 60°C for 30 s; then at 72°C for 30 s × 40 cycles. As soon as the PCR was completed, baseline and threshold values were set automatically and threshold cycle (C T) values were determined using the Biorad iCycler software (BioRad, Germany). RNA quantities were analyzed using the ΔΔ-C T method [16] using Excel sheets to calculate relative expression values.
Cloning of CCDC66 cDNAs for the analysis of differential splicing
Total RNA was isolated from canine retina using standard protocols. cDNA was subsequently generated using oligo-dt(18) nucleotides. The canine CCDC66 cDNA was amplified using the oligonucleotides detailed in supplementary Table 1. The corresponding amplicon (expected size 3.12 kb) was digested with SalI and EcoRI and cloned into the pSPT18 vector (Roche, Mannheim) using compatible restriction enzyme sites for unidirectional cloning. The construct was subsequently transfected into E. coli TOP10 electro-competent cells and plated on LB agar plates supplemented with 50 µg/ml ampicillin. The plated cells were incubated overnight, and single colonies were isolated and archived in LB (amp)-glycerol media for subsequent characterization.
Ninety-four randomly selected canine clones were characterized using eight overlapping PCR systems spanning the entire CCDC66 cDNA (supplementary Table 1). The amplicons were electrophoretically separated using 3% NuSieve agarose gels (Biozym Hessisch Oldendorf, Germany). Clones representing the various lengths as well as a full-length reference were sequenced. DNA sequencing was performed using BigDye® terminator reagents with subsequent analysis on an ABI 377 device.
Immunohistochemistry
Adult mice were deeply anesthetized with phenobarbital (720 mg/kg Nembutal i.p.) and transcardially perfused with 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4) as described [17]. The eyes were enucleated, washed in phosphate-buffered saline (PBS), and embedded in Paraffin. Eyes from four dogs (one gPRA-affected Schapendoes, two Saarloos Wolfhounds, and one Weimaraner as healthy controls) and two humans were enucleated, immersion-fixed in 4% paraformaldehyde for several hours, sagittally cut at the level of the optic nerve, and post-fixed for another 24 h. The fundus of canine and human eyes were photo-documented using the Olympus SZX-12 camera (Olympus Optical, Japan) in order to detect obvious macroscopical affections, then transferred to PBS and embedded in Paraffin. Blocks were serially cut into 15 µm sections, mounted on Superfrost Plus slides (Menzel, Braunschweig, Germany) and dried for at least 24 h at 40°C. Sections taken for immunohistochemistry were de-waxed in Xylene and rehydrated. Prior to immunohistochemical treatment, antigen retrieval was performed by incubating the sections in a solution of 10 mM citrate buffer (pH 6) for 15 min in a microwave oven (800 W) according to a modified protocol [18]. Microwaved sections were cooled down to room temperature (RT), washed in PBS and pre-incubated in a blocking solution with 20% normal goat serum (plus 0.1% Triton X-100, 0.05%phenylhydrazine, 0.01% thimerosol, and 0.1% sodium azide) diluted in PBS followed by an overnight incubation at RT with the primary CCDC66 (sc-102418; Santa Cruz Biotechnology, USA) antibody diluted in the previous solution (1:200 for mice, 1:100 for dogs and humans). Then sections were rinsed in PBS, pre-incubated with 0.2% bovine serum albumin in PBS (PBS-A) for 1 h, and incubated overnight with biotinylated goat-anti-rabbit secondary antibody (Vector Laboratories, Burlingame, CA 94010, USA) at RT. After washing with PBS, sections were incubated with the avidin-biotinylated peroxidase complex (Vector Labs) for 2 h, diluted 1:1,000 in PBS-A. Peroxidase activity was visualized with 3.3′-diaminobenzidine under microscopic control. Finally, sections were rinsed in PBS, quickly dehydrated and cover-slipped with EntellanR (MERCK KGaA, Darmstadt, Germany).
In order to control the specificity of the CCDC66 antibody, the corresponding peptide (sc-102418P; Santa Cruz) was used to eliminate immunoreactivity by prior absorption of the antibody. The antibody was mixed with a tenfold excess of the peptide (e.g. 5 µg peptide/ml in 1:200 antibody dilution) for 6 h at 4°C. Incubation proceeded as described above with the same concentration as was used for the primary antibody. Omission of primary antibody served as control for the secondary antibody.
Adjacent sections were stained with hematoxylin–eosin as morphological reference. Pictures were taken with an Olympus DP 71 camera mounted on an Olympus microscope BH-2, and documented by the computer-assisted analysis system (Soft imaging system GmbH, Münster, Germany) then exported as TIFF files into Adobe Photoshop 10.01 (Adobe Imaging Systems Inc., USA) for photo-processing.
Western blot analysis
Retina samples of adult C57BL/6 J mice were homogenized with a pre-cooled Teflon™ glass homogenizer in ice cold RIPA buffer [50 mM Tris–HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium-dodecyl sulfate (SDS) or SDS buffer (2 mM EGTA, 500 mM Tris–HCl, pH 6.8, 2% SDS)] containing protease inhibitors (P2850, Sigma-Aldrich). In order to analyze CCDC66 protein under reducing conditions, 10 mM dithiothreitol (DTT, Fermentas, St. Leon-Roth, Germany) and non-reducing conditions 8% w/v of the alkylating agent iodoacetamide (#A3221, Sigma) was added to the RIPA buffer. The homogenate was then incubated on ice and centrifuged (15 min, 1,000×g) in order to remove cell debris and nuclei. Supernatant was collected, followed by protein determination using Bradford assay (BioRad, München, Germany). Protein samples were denatured in Laemmli buffer without non-reducing conditions; or reducing conditions containing beta-mercaptoethanol (0.5%, 1%, or 2%); or 100 mM Dithiothreitol (Fermentas, St. Leon-Roth, Germany), and resolved together with a protein molecular weight marker (#161-0375, BioRad; #SM1851, Fermentas) on 10% SDS gels. Electrophoretic transfer onto nitrocellulose (Hybond C 0.8 µm pore size, GE Healthcare, Munich, Germany) was performed in a tank-blot system using transfer buffer (48 mM Tris, 39 mM Glycin, 0.375 % SDS, 20% methanol) at 30 V and 4°C overnight. Membrane bound proteins were visualized by staining with ponceau S, de-stained and unspecific binding sites were blocked by incubation in PBS-blocking solution (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, and 2.5% non-fat dry milk, pH 7.4). The primary antibody against CCDC66 antigen (dilution 1:250, G-14 antibody #sc-102418, Santa Cruz, USA) was incubated in PBS/1.25% non-fat dry milk at 4°C overnight. After six washes in PBS-Tween 0.1% the secondary antibody (dilution, 1:5,000, #111-035-003, HRP-conjugated goat-anti-rabbit antibody, Jackson Immuno Research, London) was incubated in PBS/1.25% non-fat dry milk for 1 h at RT followed by six washes in PBS–Tween 0.1%. In order to compare the applied protein amounts in the different lanes, antibodies were stripped at 37°C (60 mM Tris, pH 6.7; 2% SDS, 100 mM β-Mercaptoethanol), incubated again in PBS-blocking solution at 4°C overnight followed by incubation in β-actin antibody (dilution 1:5,000, AC-15 antibody # A1978, Sigma) and wash in PBS-Tween 0.1%. This was followed by incubation with secondary antibody (dilution 1:7,500, #315-035-045, HRP-conjugated rabbit-anti-mouse antibody, Jackson Immuno Research, London) and final wash in PBS-Tween. Following the manufacturer’s instructions, bound antibodies were detected using a chemiluminescence kit (ECL plus, GE Healthcare) and a laser scanner documentation system (Storm 860, GE Healthcare).
Results
Fine mapping and candidate gene analysis
Twelve gPRA affected, four carrier, and one healthy Schapendoes from two informative pedigrees with segregating gPRA were genotyped with eight CFA20 microsatellites in the region between the markers FH3358 and TL336MS spanning 5.6 Mb [9]. This candidate region was subsequently reduced to 4.2 Mb by virtue of the affected haplotype information based on additionally identified recombination events (Fig. 1). The homologous region on human chromosome 3p23.1 contains 10 candidate genes out of which eight genes are expressed in the human retina to varying degree (UCSC Genome Browser).
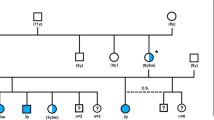
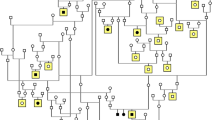
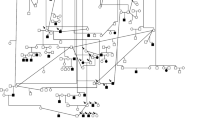
The CFA 20 candidate region for gPRA in Schapendoes including all candidate genes analyzed, the newly identified CCDC66 mutation and haplotype segregation analyses demonstrating recombination events. a Physical map of the investigated candidate genes in the gPRA critical region comprising 4.2 Mb of DNA; b exon/intron structure of the newly identified CCDC66 gene (database entry XM_533786) in addition to the UCSC database entry herein exons 1–4, 6, 7, and 19 are demonstrated as well as additional 3′-part of exon 20 as identified by cDNA investigation. c Chromatograms of part of CCDC66 exon 6 exhibit the wild-type sequence and an A insertion in homozygous state leading immediately to a stop codon in affected dogs; d comparison of the protein sequences of exon 6 of the CCDC66 gene in eight different mammalian species; e haplotypes in dog pedigrees used for linkage analysis ([9]) in order to identify recombinations of the indirect diagnostic marker (REN93E07) and the insertion mutation in the CCDC66 gene. Mu mutation, wt wild type; circle female, square male dogs, white unaffected, black affected, white/black carrier, shaded dogs with identified recombinations
For mutation analysis in the candidate genes, five gPRA affected, five carrier, and five healthy Schapendoes were selected from the respective informative pedigrees (see [9]). Polymorphisms were identified by SSCP analysis in the ARF4, RHGEF63, and C3ORF63 genes (Table 1). Derived by automated computational analysis, long-range PCR spanning a gap in the reported canine CCDC66 sequence identified exons 6 and 7, which are not included in the published CCDC66 gene (XM_533786).
In the newly identified sequence of exon 6 (see Fig. 1d for trans-species comparison); SSCP analysis and subsequent direct sequencing revealed an inserted base A (c.521_522InsA) in a homozygous state in the five affected Schapendoes. This insertion was subsequently verified in the remaining 14 affected Schapendoes. The mutation results in a frame shift that directly leads to a stop codon and to in silico translation of a truncated protein (p.Asn174LysfsX) in affected gPRA dogs. The insertion mutation had not been observed in 50 additional gPRA-affected dogs from 29 different breeds and 41 healthy dogs from 15 different breeds.
Exon 6 of the CCDC66 gene was subsequently investigated by DHPLC in 870 Schapendoes (including 19 affected dogs and 156 carriers) that had previously undergone indirect diagnostic testing for gPRA. In 150 of the 156 dogs (96.15%) that were classified as potential carriers by the indirect test, the CCDC66 mutation was present in the heterozygous state. The six dogs classified as carriers before (3.85%) were homozygous for the wild-type allele, suggesting that recombination must have occurred between the marker for the indirect diagnostic test and the mutation. In order to identify the breakpoint(s) in these distantly related dogs, we performed additional haplotype analyses. These investigations revealed that different recombination events have happened in different families; one between markers FK37.8_20 and REN316E23, another one between FK37.3_20 and FK37.8_20 (see Fig. 1). In these regions, Alu elements are present which contain short tandem repeat stretches.
Characterization of the CCDC66 gene
Until now, the CCDC66 gene had never been characterized in detail in either the human or canine genomes. The structure and evolutionary conservation of CCDC66 was investigated here by alignments of human and mouse genomic DNA, cDNA, and protein sequences. Comparison of human cDNAs [15] with sequences of the dog database provided limited information on the intron/exon boundaries for mutation analyses. Since gaps were still present in the canine genomic CCDC66 sequence as provided in the UCSC database, the respective regions were amplified by long-range PCR and sequenced. Comparisons of the canine CCDC66 gene with human and mouse sequences showed overall nucleotide conservation rates of 66–82% for exon 6 (dog/man 82%; dog/mouse 69%; man/mouse 66%) and 91–92% for exon 7 (dog/man and dog/mouse 91%; man/mouse 92%). Overlapping PCR with cDNAs verified the newly identified exons in dogs. Long-range PCR products of cDNAs spanning exon 6 and the 3′UTR showed only minor variations, whereas the amplification products including exon 6 to the 5′ region exhibited high variability (for further details, see below). The human CCDC66 gene is located on chromosome 3 with 19 exons encompassing ∼64.6 kb, whereas the canine homologue is located on CFA20 comprising 20 exons and spanning ∼44.7 kb. Exon lengths vary between man and dog by up to 45 bp. A homologous sequence to canine exon 9 has not yet been annotated in human cDNAs. A variety of open reading frames have been identified in man, dog, and mouse arising from various in frame start codons and alternative splicing (Fig. 2). The predicted open reading frame (ORF) in man encodes a 914 aa protein (NM_001012506). The corresponding ORF in dogs encodes a protein of 917 amino acids (aa) and in mice, 935 aa. According to ECgene analysis, a clustering summary for H3C7774 (alias CCDC66) with medium confidence level (B) produces 20 transcript variations encoding 12 distinct proteins (compare Fig. 2). The EST database (www.ncbi.nlm.nih.gov/UniGene) implies that the CCDC66 gene is expressed in a wide range of tissues.
The expression of CCDC66 RNA varies physiologically in different mouse tissues (see Fig. 3a). This RT-PCR was performed with primers hybridizing to sequences in exons 5 and 6, thus always encompassing the homologous sequence corresponding to the Schapendoes’ gPRA mutation in exon 6. The retina-specific GNAT1 and the ubiquitously expressed HPRT genes served as controls in the expression studies. In order to evaluate the consequences of the insertion mutation, we analyzed CCDC66 expression in the retinae and blood of affected as well as unaffected Schapendoes. Using RT-PCR, cDNA was amplified corresponding to genomic sequences located 5′ of the mutation in exon 6. The CCDC66 RNA expression is detectable at very low levels in the retina of the affected Schapendoes, but to a much lower degree than in unaffected dogs (Fig. 3). The retina-specific GNAT1 gene is not expressed in blood and, as expected, highly expressed in retinae of unaffected dogs.
Quantitative real-time RT-PCR of total RNA extracted from mouse (a) and canine (b) tissues using different amplification systems for mouse and dog. The relative expression of CCDC66, HPRT, and GNAT1 is normalized to the amount of GAPDH in the same cDNA sample. Retinal CCDC66 expression is set to 1, and all values are depicted in relation to the latter. The standard deviation (indicated by error bars) is calculated from a set of two independent experiments. a CCDC66 RNA shows similar expression levels in the selected mouse tissues, except for very low expression in muscle. b Retinal CCDC66 expression correlates with the expression of the rod-photoreceptor specific gene GNAT1 in the healthy and gPRA-affected retina. Retinal degradation of the gPRA-affected retina also reveals an increased expression of the apoptosis marker HPRT. Note that for the canine CCDC66 expression studies the employed RT-PCR system amplifies only a portion of the cDNA located 5′ of the mutation
Differential splicing products
Ninety-four randomly selected canine CCDC66 cDNA clones revealed 17 different splice variations (Fig. 2). The most common form (45%) comprises 917 in silico translated aa with a predicted molecular weight of 106 kDa. The respective translation initiation site is homologous to the one reported in human CCDC66 (UniProtKB/Swiss-Prot A2RUB6). The second most frequent isoform (24%) differs from the more frequent one by a different amino-terminal methionine. Comparison of putative ATG translation initiation sites for these two variations with the classical Kozak motif [19] revealed two ATG codons to represent the translation initiation sites with high probability.
The remaining 15 variants sum up to 31% with frequencies between 1–5%. These rarer isoforms exhibit high variability 5′ and 3′ of the detected insertion mutation including deletions of entire exons as well as insertions of additional intronic sequences, leading in silico to truncated or abbreviated proteins. Furthermore, sequence analysis of the cDNAs showing 3′ variability revealed incomplete splicing or intermediary products as indicated by persisting splice donor and acceptor site motifs (GT/AG). Thus, as expected, unselective cDNA generation from RNA using oligo-dt(18) primer and cloning of all CCDC66 transcripts yields also immature or incompletely spliced products respectively. Hence, there is evidence for two common cDNA varieties that encode functional canine CCDC66 protein isoforms.
CCDC66 protein in the retina of mouse, dog and man
Immunohistochemistry demonstrated CCDC66 reaction product in the retinae of mouse, dog, and man in a similar distribution pattern (Fig. 4). All healthy specimen investigated showed the strongest CCDC66 staining in the inner segments of the photoreceptors (Fig. 4a, d, g), independent of the species’ origin. Faint immunoreactivity was also detectable in the outer segments, the outer plexiform and the ganglion cell layers with minor species-dependent differences in expression. Immunoreactivity was eliminated by pre-absorption of the CCDC66 antibody with the corresponding peptide, confirming the high specificity of the antibody used (Fig. 4b, e, h). The normal, unaffected structure of all retinal layers was documented by adjacent hematoxylin–eosin-stained sections (Fig. 4c, f, i).
Micrographs of vertical 15-µm thick paraffin sections of mouse (a–c), human (d–f), and dog retinae (g–i). Sections stained for CCDC66 show strong immunoreactivity in the inner segments (IS) of the photoreceptor layer of all species investigated (a, d, g). Faint immunoreaction is observed in the outer segments (OS), outer plexiform layer (OPL), and ganglion cell layer (GCL). Control sections exhibit no immunoreactivity (b, e, h). Hematoxylin–eosin (HE) staining of adjacent sections shows the normal retinal layers (c, f, i). (k) Fundoscopy of a gPRA-affected Schapendoes eye exhibits irregular pigment distribution with some larger spots (arrows). l CCDC66-immunostained paraffin section of the peripheral retina lacks immunoreactivity. Single pigment deposits (x) are also detected in the adjacent HE-stained section (m) showing that retinal layers have disappeared. n Fundoscopy including the optic disk (OD) displays hyper-reflectivity and thinning of retinal vessels. o Paraffin section of the central retina immunostained for CCDC66 also lacks specific reaction in all layers including the IS (arrows), which were identified in the adjacent HE-stained section (p). Note the thin outer nuclear layer (ONL) and shortened IS (arrows). Inner nuclear layer (INL), inner plexiform layer (IPL). Scale bars represent 20 µm in b for a–c, in e for d–f, in h for g–i, and in o for l, m, o, p
The retina of a gPRA-affected, 6-year-old Schapendoes displayed all the signs of advanced retinal degeneration. Fundoscopy showed irregularly, pigmented areas with dark spots (Fig. 4k) also detected as single or confluent pigment deposits in paraffin sections of the severely degenerated peripheral parts (Fig. 4l, m). There, the regular retinal layers are lost completely. These sections immunolabeled for CCDC66 exhibited no specific reactivity (Fig. 4l). Fundoscopy, including the optic disk, showed marked attenuation of vessel diameter and hyper-reflectivity of the tapetum as ophthalmoscopic hints of retinal affection (Fig. 4n). The central area around the optic disk displayed a very small isolated field with retinal layers preserved, albeit the outer nuclear layer was reduced in thickness and the inner segments shortened (Fig. 4p). Immunostaining of adjacent sections (Fig. 4o) confirmed that the inner segments, the outer plexiform, and the ganglion cell layers lacked CCDC66 protein in the retina of this gPRA-affected Schapendoes, which was homozygous for the newly identified mutation described above.
In Western blots, CCDC66 protein expression was detected in the mouse retina and the polyclonal antibody revealed a strong signal band at 220 kDa; corresponding to the predicted size of dimeric CCDC66 with an ORF of 949 amino acids (Fig. 5). The antibody consistently detected another protein of ∼37 kDa, possibly representing a protein from an alternative splice product. Because of the probability that highly reactive thiol groups in the CCDC66 protein may react and form dimers in the presence of 0.5% β-Mercaptoethanol, CCDC66 dimer formation was studied under varied conditions (Fig. 5b). After alkylation of free SH groups with iodoacetamide under non-reducing conditions, a strong dimer band of about 220 kDa indicated that dimer formation is not the result of disulfide bonding during lysis, preparation, or electrophoresis. In contrast, dimers are affected by increasing β-mercaptoethanol concentrations as well as by reducing conditions using dithiothreitol, revealing a monomer band at the predicted molecular weight of 110 kDa. Altogether, this approach revealed that a high proportion of CCDC66 protein exists as dimers that are sensitive to reducing conditions either by presence of β-mercaptoethanol or dithiothreitol. This fact implies that dimerization involves disulfide bonds.
Immunoblot analysis of CCDC66 protein expression in mouse retina. a An intense signal (∼230 kDa) and a faint band (37 kDa; bottom asterisk) are detected with the CCDC66 antibody. Because the predicted molecular masses for the described isoforms of CCDC66 do not exceed ∼109 kDa, the high molecular weight band (top asterisk) may represent dimers. A smaller isoform with an apparent molecular weight of ∼35 kDa is present in mouse retina. This corresponds to a predicted human isoform (Uniprot accession number A2RUB6 isoforms are: ∼109, ∼105, 32, and 21 kDa). SDS homogenization buffer, 0.5% ß-mercaptoethanol, 40 µg protein/lane. b Under reducing conditions with ß-mercaptoethanol or DTT the 110 kDa isoform of CCDC66 protein predominates, the proposed dimer staining intensity decreases. In contrast, dimer formation predominates after alkylation of SH groups using iodoacetamide. D dimer, DTT dithiothereitiol (100 mM), IAA iodoacetamide (8% w/v), M monomer, ßMe ß-Mercaptoethanol, R RIPA homogenization buffer, S SDS homogenization buffer. c Similar protein amounts (40 µg) have been loaded into the different lanes as can be evidenced by β-actin antibody staining
Discussion
A CCDC66 insertion mutation in the homozygous state is associated with gPRA in Schapendoes; resulting in a premature stop codon and abrogated protein translation. The in silico deduced polypeptide is truncated and thus, probably, it will be degraded quickly. The relevance of the newly identified CCDC66 gene is paramount as evidenced by high sequence conservation in vertebrates. CCDC66 mRNAs are differentially spliced. After cloning of the full-length cDNA, 17 splice variations are obvious; five of which lead theoretically to proteins with different ORFs. Expression studies in dogs and mice via qRT-PCR showed that CCDC66 RNA is present in many tissues. In the retina, CCDC66 protein expression is prominent in the inner segments of photoreceptor cells and at a lower level in the external plexiform and ganglion cell layers in mouse, dog, and man. In affected Schapendoes, decreased CCDC66 RNA levels are obvious in retinal remnants (as demonstrated also for retina-specific GNAT1 RNA). Thus, reduced CCDC66 expression is mainly dependent on the photoreceptor cells that are continuously lost during the course of gPRA.
RNA expression of CCDC66 gene in adult mouse tissue indicates that transcription is not restricted to the retina. Are dogs characterized by similar expression patterns? How can widespread expression be reconciled with the fact that no other symptoms appear associated with CCDC66 deficiency except for PRA? In man, a few genes are widely expressed in multiple adult tissues but only cause disease pathology within the retina and result in the human counterpart of PRA, i.e. RP. These genes are known to be involved in autosomal dominant [20–22], X-linked [23, 24] and also in autosomal recessive RP forms. Tuson et al. [25] report two families with mutations in the CERKL gene, encoding a ceramide kinase expressed in many tissues, leading to a premature termination codon in the protein product. The pathomechanism is suspected to be due to enzyme deficiency, resulting in metabolite overrun and sphingolipid-mediated apoptosis of retinal cells. Moreover, Hartong et al. [26] identified two families with RP due to mutations in IDH3B, a gene encoding an enzyme of the citric acid cycle. In individuals with IDH3B mutations in homozygous state the defect may be substituted by other enzymes in all tissues except for the retina. These studies indicate that gene expression in multiple tissues is able to result in retinal degeneration only. Nevertheless, based on our findings that various CCDC66 splice variations in the canine retina exist as well as different protein isoforms, a retina-specific transcript could result in restricted damage to the retina. Retina-specifically spliced variations and/or protein isoforms would then account for the healthy phenotype in Schapendoes except for PRA.
CCDC66 proteins have a heptad repeat pattern which contains at least one coiled-coil domain; a secondary structure composed of two or more α-helices which form a cable-like structure. In proteins, the helical cables serve a mechanical role by forming stiff bundles of fibers [27]. Coiled-coil domains were discovered first in α-keratin [28], and they occur as structural elements in many other proteins. Coiled-coil proteins usually interact with other coiled-coil proteins and participate in a plethora of protein–protein interactions [29]. Our initial results indicate that a large part of the CCDC66 protein is indeed present in dimers. Hence, we concentrate on the biochemical characterization of CCDC66 and interacting proteins in ongoing studies. In addition, CCDC66 expression during retinal development remains so far an entirely open issue that may render crucial insights into functional aspects of this protein. Currently, the CCDC66 gene is also screened for mutations in familial cases of RP. Even if no mutation would be detected in the DNAs from RP patients, studies on the (patho)physiology of the newly identified protein and its potential molecular interaction partners may yield crucial insights into additional RP forms like it has already been elaborated e.g. for RPE65, an isomerohydrolase expressed in retinal pigment epithelium. For the latter defect specific gene therapy approaches have been devised [30].
In conclusion, we submit that a mutation in the CCDC66 gene causes gPRA in Schapendoes. This argument is strengthened with the fact of specific protein expression in photoreceptor cells. Thus, the stage is set to study the pathogenesis of gPRA based on lacking CCDC66 protein as well as its natural function(s) in vivo.
References
Petersen-Jones S (2005) Advances in the molecular understanding of canine retinal diseases. J Small Anim Pract 46:371–380
Dekomien G, Runte M, Godde R, Epplen JT (2000) Generalized progressive retinal atrophy of Sloughi dogs is due to an 8-bp insertion in exon 21 of the PDE6B gene. Cytogenet Cell Genet 90:261–267
Goldstein O, Zangerl B, Pearce-Kelling S, Sidjanin DJ, Kijas JW, Felix J et al (2006) Linkage disequilibrium mapping in domestic dog breeds narrows the progressive rod-cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics 88:541–550
Mellersh CS, Boursnell ME, Pettitt L, Ryder EJ, Holmes NG, Grafham D et al (2006) Canine RPGRIP1 mutation establishes cone-rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics 88:293–301
Petersen-Jones SM, Entz DD, Sargan DR (1999) cGMP phosphodiesterase-alpha mutation causes progressive retinal atrophy in the Cardigan Welsh corgi dog. Invest Ophthalmol Vis Sci 40:1637–1644
Suber ML, Pittler SJ, Qin N, Wright GC, Holcombe V, Lee RH et al (1993) Irish setter dogs affected with rod/cone dysplasia contain a nonsense mutation in the rod cGMP phosphodiesterase beta-subunit gene. Proc Natl Acad Sci U S A 90:3968–3972
Zangerl B, Goldstein O, Philp AR, Lindauer SJ, Pearce-Kelling SE, Mullins RF et al (2006) Identical mutation in a novel retinal gene causes progressive rod-cone degeneration in dogs and retinitis pigmentosa in humans. Genomics 88:551–563
Zhang Q, Acland GM, Parshall CJ, Haskell J, Ray K, Aguirre GD (1998) Characterization of canine photoreceptor phosducin cDNA and identification of a sequence variant in dogs with photoreceptor dysplasia. Gene 215:231–239
Lippmann T, Jonkisz A, Dobosz T, Petrasch-Parwez E, Epplen JT, Dekomien G (2007) Haplotype-defined linkage region for gPRA in Schapendoes dogs. Mol Vis 13:174–180
Lippmann T, Pasternack SM, Kraczyk B, Dudek SE, Dekomien G (2006) Indirect exclusion of four candidate genes for generalized progressive retinal atrophy in several breeds of dogs. J Negat Results Biomed 5:19
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Jaeckel S, Epplen JT, Kauth M, Miterski B, Tschentscher F, Epplen C (1998) Polymerase chain reaction-single strand conformation polymorphism or how to detect reliably and efficiently each sequence variation in many samples and many genes. Electrophoresis 19:3055–3061
Bennett RR, den Dunnen J, O'Brien KF, Darras BT, Kunkel LM (2001) Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet 2:17
Xiao W, Oefner PJ (2001) Denaturing high-performance liquid chromatography: a review. Hum Mutat 17:439–474
Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R et al (2004) Complete sequencing and characterization of 21, 243 full-length human cDNAs. Nat Genet 36:40–45
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Petrasch-Parwez E, Habbes HW, Weickert S, Lobbecke-Schumacher M, Striedinger K, Wieczorek S et al (2004) Fine-structural analysis and connexin expression in the retina of a transgenic model of Huntington's disease. J Comp Neurol 479:181–197
Fricke B, Parsons SF, Knopfle G, von During M, Stewart GW (2005) Stomatin is mis-trafficked in the erythrocytes of overhydrated hereditary stomatocytosis, and is absent from normal primitive yolk sac-derived erythrocytes. Br J Haematol 131:265–277
Kozak M (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 12:857–872
Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C et al (2001) A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8:375–381
McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ et al (2001) Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 10:1555–1562
Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG et al (2002) Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet 11:87–92
Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G et al (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332
Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A et al (1996) A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X- inked retinitis pigmentosa (RP3). Nat Genet 13:35–42
Tuson M, Marfany G, Gonzàlez-Duarte R (2004) Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26). Am J Hum Genet 74:128–138
Hartong DT, Dange M, McGee TL, Berson EL, Dryja TP, Colman RF (2008) Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet 40:1230–1234
Liu J, Zheng Q, Deng Y, Cheng CS, Kallenbach NR, Lu M (2006) A seven-helix coiled coil. Proc Natl Acad Sci U S A 103:15457–15462
Crick FH (1952) Is alpha-keratin a coiled coil? Nature 170:882–883
Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88
Cai X, Conley SM, Naash MI (2009) RPE65: role in the visual cycle, human retinal disease, and gene therapy. Ophthalmic Genet 30:57–62
Acknowledgements
We thank the owners of the dogs for blood samples, especially H. Mohr and G. de Wit-Bazelmans for their support; the veterinarians of the Dortmunder Ophthalmologenkreis (DOK; Dr. R. Brahm) for the ophthalmologic examinations and enucleation; J. Rutten who carried out parts of the fine mapping and the analysis of candidate genes; and S. Oberland who investigated the C3ORF63 gene. We thank K. Rumpf, A. Schlichting, and H.-W. Habbes for their excellent technical assistance and B. Manderson for proofreading. These studies were supported in part by the Gesellschaft für kynologische Forschung (Bonn, Germany).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
a: Oligonucleotides used for mutation screening, DNA sequencing, splicing, cloning and qRT-PCR analyses in dog and mouse. *#: Oligonucleotides were also used for splice analysis (DOC 352 kb)
Supplementary Table 2
b: Oligonucleotides used for linkage analysis and fine mapping of the candidate region. F primers were tailed using the unique sequence CAT CGC TGA TTC GCA for HEX or FAM labeling needed for genotyping analyses (DOC 60 kb)
Rights and permissions
About this article
Cite this article
Dekomien, G., Vollrath, C., Petrasch-Parwez, E. et al. Progressive retinal atrophy in Schapendoes dogs: mutation of the newly identified CCDC66 gene. Neurogenetics 11, 163–174 (2010). https://doi.org/10.1007/s10048-009-0223-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-009-0223-z