Abstract
Mast syndrome (SPG21) is an autosomal-recessive complicated form of hereditary spastic paraplegia characterized by dementia, thin corpus callosum, white matter abnormalities, and cerebellar and extrapyramidal signs in addition to spastic paraparesis. A nucleotide insertion resulting in premature truncation of the SPG21 gene product acidic cluster protein 33 (ACP33)/maspardin underlies this disorder, likely causing loss of protein function. However, little is known about the function of maspardin. Here, we report that maspardin localizes prominently to cytoplasm as well as to membranes, possibly at trans-Golgi network/late endosomal compartments. Immunoprecipitation of maspardin with identification of coprecipitating proteins by mass spectrometry revealed the aldehyde dehydrogenase ALDH16A1 as an interacting protein. This interaction was confirmed using overexpressed proteins as well as by fusion protein pull down experiments, and these proteins colocalized in cells. Further studies of the function of ALDH16A1 and the role of the maspardin–ALDH16A1 interaction in neuronal cells may clarify the cellular pathogenesis of Mast syndrome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The hereditary spastic paraplegias (HSPs) are a group of neurological disorders characterized by progressive spasticity and weakness of the lower limbs [1–8]. They are classified as “pure” if lower extremity spasticity and weakness occur in isolation or “complicated” if patients exhibit other neurological abnormalities such as cognitive impairment, ataxia, distal amyotrophy, or thinning of the corpus callosum [2]. The HSPs characteristically result from axonal degeneration in the distal portions of long descending corticospinal tracts and ascending dorsal column fibers, which are among the longest motor and sensory axons, respectively, in the central nervous system [6]. More recently, the identification of multiple genetic loci (SPG1-40) for the HSPs has permitted a molecular classification of these disorders [3–8]. Seventeen disease genes for HSPs have now been identified, and based on the proteins identified several mechanisms for pathogenesis have been proposed. These include abnormal cell signaling or migration, mitochondrial abnormalities, aberrations of myelination, impaired cholesterol or neurosteroid metabolism, and deficiencies in intracellular trafficking and transport. Most known proteins appear to fall into the latter category, including atlastin-1 (SPG3A), spastin (SPG4), NIPA1 (SPG6), strumpellin (SPG8), KIF5A (SPG10), spastizin (SPG15), seipin (SPG17), spartin (SPG20), acidic cluster protein 33 (ACP33)/maspardin (SPG21), REEP1 (SPG31), and ZFYVE27/protrudin (SPG33) [3–8].
Mast syndrome (SPG21; MIM 248900) is a childhood-onset, autosomal-recessive, complicated HSP in which progressive spastic paraparesis is associated with other abnormalities including dementia, developmental defects, a thin corpus callosum, white matter abnormalities, and cerebellar and extrapyramidal signs [9]. The relatively early onset of SPG21 has led to suggestions that it may be at least in part a neurodevelopmental disorder, though it is clearly progressive during adulthood. SPG21 is caused by a nucleotide insertion (601insA) in exon 7 of the ACP33/maspardin gene, resulting in a frame shift and subsequent premature termination (fs201-212X213) of the 308-amino-acid maspardin protein [9]. Thus, disease pathogenesis is likely due to a lack of functional maspardin protein.
The ACP33/maspardin protein harbors a single known domain with limited similarity to the α/β hydrolase superfamily. In ACP33/maspardin, this domain comprises a nucleophile elbow and parallel β strands, but it lacks the catalytic triad common to most α/β hydrolases, suggesting that it lacks enzymatic activity and might instead function as a peptide-binding module mediating protein–protein interactions [9]. In fact, ACP33/maspardin was originally identified as an intracellular binding protein for the cell surface glycoprotein CD4 and proposed to modulate CD4 stimulatory activity [10]. Within the nucleophile elbow of the α/β hydrolase domain, Ser109 is critical for the interaction of maspardin with CD4 [10]. However, ACP33/maspardin is widely expressed in many cell types other than T cells, including cells lacking CD4 such as neurons. Therefore, ACP33/maspardin likely also interacts with other proteins.
In this study, we examined the localization of ACP33/maspardin using immunocytochemistry and biochemical fractionation. We found that ACP33/maspardin localizes to both cytoplasm and at membranes of the endosomal/trans-Golgi network. An immunoprecipitation screen of HeLa cell lysates using an anti-ACP33/maspardin antibody identified the aldehyde dehydrogenase ALDH16A1 as an endogenous interacting protein that colocalizes partially with ACP33/maspardin in both neurons and nonneuronal cells.
Materials and methods
DNA constructs and cell culture
The human maspardin complementary DNA (cDNA; GenBank accession no. NM_016630) was cloned into the EcoRI site of the mammalian expression vector pGW1 with an N-terminal Myc or HA-epitope tag [11, 12] as well as the EcoRI site of pRK5 (Genentech, South San Francisco, CA, USA) for expression of untagged protein. Additionally, full-length maspardin was cloned into the EcoRI site of the bacterial fusion protein vector pGEX-6P-1 (GE Healthcare Life Sciences, Piscataway, NJ, USA). The full-length human ALDH16A1 (GenBank accession no. NM_153329) cDNA was cloned into the EcoRI site of pGW1. In addition, ALDH16A1 was cloned into the EcoRI site of pCAL-n-EK (Stratagene, La Jolla, CA, USA) for production of calmodulin-binding peptide (CBP)-ALDH16A1 bacterial fusion proteins. Site-directed mutagenesis was performed using the QuikChange method (Stratagene).
Antibodies
Two rabbit polyclonal anti-peptide antibodies against human maspardin were generated and affinity-purified commercially (BioSource International, Hopkinton, MA, USA); one targeted N-terminal amino acid residues 1–18 (b5704; acetyl-MGEIKVSPDYNWFRGTVPC-amide), and the other was raised against C-terminal residues 283–300 (c3945; acetyl-CAIDPSMVSAEELEVQKGS-amide). Additionally, an anti-maspardin polyclonal antibody (IgY) raised in chickens was purchased from GenWay Biotech (San Diego, CA, USA). Rabbit polyclonal anti-ALDH16A1 antibodies were produced and affinity-purified commercially against a peptide corresponding to residues 307–324 (#5650; acetyl-CLRLLIQESVWDEAMRRLQ-amide; BioSource International). Mouse monoclonal anti-PLCγ-1 (B-6-4) antibodies were obtained from Millipore (Billerica, MA, USA). Mouse monoclonal antibodies against Lamin A + C (131C3; IgG1) and mannose-6-phosphate receptor (2G11; IgG2a) as well as rabbit polyclonal antibodies against ALDH1A1 were purchased from Abcam (Cambridge, MA, USA). Mouse monoclonal anti-GM130 (clone 35; IgG1), anti-EEA1 (clone 14; IgG1), anti-CD63 (H5C6; IgG1), and anti-OPA1 (clone 18; IgG1) antibodies were from BD Biosciences (San Jose, CA, USA). Mouse monoclonal anti-γ-adaptin (100/3; IgG2b) antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse monoclonal anti-LAMP1 (H4A3; IgG1) was from SouthernBiotech (Birmingham, AL, USA). Mouse monoclonal anti-Myc (9E10; IgG1) and anti-HA probes (F-7; IgG2a) and rabbit polyclonal anti-calnexin (H-70) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture and immunofluorescence microscopy
Human adenocarcinoma (HeLa) cells (American Type Culture Collection CCL-2) were maintained, transfected, and harvested as described previously [11–13]. Primary cultures of mouse cerebral cortical neurons were prepared from P0 mice, plated at a density of ∼1.0 × 104 per square centimeter on coverslips, and maintained as described previously [14]. At 4 days in vitro (DIV4), neurons were fixed and stained as described below for HeLa cells. All animal experiments were approved by the National Institute of Neurological Disorders and Stroke (NINDS)/National Institute on Deafness and Other Communication Disorders Animal Care and Use Committee.
Approximately 24 h after plating, HeLa cells were washed with phosphate-buffered saline (PBS; pH 7.4), fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, and washed again with PBS. Cells were then permeabilized and incubated with blocking solution (40% horse serum, 0.1% bovine serum albumin, and 0.1% Triton X-100 in PBS) for 1 h and subsequently with rabbit polyclonal anti-maspardin (9 μg/ml) antibodies in blocking solution for 1 h at room temperature. After washing with PBS, cells were incubated with anti-rabbit Alexa-Fluor-568-conjugated secondary antibodies (1:500; Invitrogen, Carlsbad, CA, USA) in blocking solution for 30 min at room temperature and washed again with PBS. Specificity of immunostaining was determined by comparisons with antibodies preincubated with antigenic peptide (1 μM) for 1 h at 4°C. For colocalization experiments with organelle-specific markers, anti-maspardin and secondary antibody concentrations remained the same. Each monoclonal antibody (anti-EEA1 [2.5 μg/ml], anti-GM130 [2.5 μg/ml], anti-γ-adaptin [1:100], anti-mannose-6-phophate receptor [2.0 μg/ml], anti-LAMP1 [1.0 μg/ml], or anti-CD63 [0.5 μg/ml]) was applied to coverslips and visualized with anti-mouse Alexa-Fluor-488-conjugated secondary antibodies (1:1,000) in conjunction with anti-maspardin antibodies. To visualize maspardin and ALDH16A1 colocalization, chicken polyclonal anti-maspardin (5 μg/ml) and its appropriate secondary (goat anti-chicken Alexa Fluor 488; 1:1,000) antibody and rabbit polyclonal anti-ALDH16A1 (15 μg/ml) and its secondary antibody (goat anti-rabbit Alexa Fluor 568; 1:1,000) were utilized. In all cases, coverslips were mounted using Gel/Mount reagent (Biomeda, Foster City, CA, USA). Fluorescence images were acquired using a Zeiss LSM-510 confocal microscope and processed with Adobe Photoshop 7.0 software.
Protein preparation, gel electrophoresis, and immunoblotting
HeLa cells were washed twice with PBS and then harvested in 0.5% Triton X-100/PBS. After multiple passages through a 25-gauge needle, the homogenate was centrifuged at 1,330×g for 3 min and subsequently at 21,000×g for 30 min to remove nuclear and cellular debris. For immunoblotting, proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electrophoretically transferred to nitrocellulose. After blocking with 5% nonfat milk/0.1% Tween 20/Tris-buffered saline (TBS; pH 7.4), antibodies (1–5 μg/ml) were added for 2.5 h at room temperature. Following several washes in TBS, horseradish-peroxidase-conjugated secondary antibodies (1:1,000; GE Healthcare Life Sciences) were added for 1.5 h in blocking buffer. Finally, after several washes with 0.1% Tween/TBS, immunoreactive proteins were revealed using Renaissance Enhanced Chemiluminescence Reagent (PerkinElmer Life Sciences, Waltham, MA, USA). Images were acquired using the Molecular Imager ChemiDoc XRS System and Quantity One software (Bio-Rad, Hercules, CA, USA).
Subcellular fractionation
Subcellular fractionation of HeLa cells was performed using the Qproteome kit (Qiagen, Valencia, CA, USA). Equal proportions of fraction lysates were resolved by SDS-PAGE and immunoblotted as described previously. In other experiments, HeLa cells were harvested in ice-cold hypotonic buffer (10 mM Tris–HCl [pH 7.4], 10 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 10 mM sodium fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride) and then disrupted using a Dounce homogenizer (50 strokes). The homogenate was centrifuged (1,330×g, 3 min, 4°C), generating a pellet (P1) and a supernatant (S1). The S1 supernatant was centrifuged at 21,200×g (10 min, 4°C), producing a pellet (P2) and a supernatant (S2). The S2 supernatant was then centrifuged at 200,000×g for 1 h at 4°C, generating a P3 pellet and S3 supernatant.
Gel-exclusion FPLC
HeLa cells were lysed in 0.1% Triton X-100/PBS and clarified by centrifugation at 130,000×g for 30 min. The soluble extract was applied to a Superdex 200 HR10/30 fast protein liquid chromatography (FPLC) column at a flow rate of 0.4 ml/min in 0.1% Triton X-100/PBS and controlled with Unicorn 5.10 software (AKTA FPLC system; GE Healthcare Life Sciences). Fractions (0.5 ml) were collected and then immunoblotted using anti-maspardin antibodies as described above. Protein standards in 0.1% Triton X-100/PBS were applied to the column to generate a standard curve, from which molecular masses were calculated.
Immunoprecipitations
HeLa cells were washed twice with PBS, harvested in 0.5% Triton X-100/PBS, and passed several times through a 25-gauge needle. Extracts were then clarified by serial centrifugations of 1,300×g for 3 min, 21,000×g for 30 min, and 100,000×g for 30 min at 4°C. Extracts (100 μg of protein) were precleared with protein A–Sepharose (Sigma-Aldrich) for 1 h and then divided equally into three samples. One sample was incubated with 5 μg of rabbit polyclonal anti-maspardin probe antibodies (c3945) and another with 5 μg nonimmune rabbit IgG for 1–2 h; then, both were subsequently incubated with protein A–Sepharose for 1 h at 4°C. The last aliquot contained protein A–Sepharose beads alone. Beads were washed three times with 0.5% Triton X-100/PBS. Bound proteins were resolved by SDS-PAGE, and proteins were visualized by staining using Coomassie Brilliant Blue. Specific bands were excised for matrix-assisted laser desorption/ionization (MALDI)–time-of-flight (TOF) mass spectrometry. Alternatively, HeLa cells transfected with HA- or Myc-ALDH16A1 were washed and extracts prepared as described above. Extracts (100 μg of protein) were incubated with rabbit polyclonal anti-maspardin antibodies (c3945 or b5704) and subsequently incubated with protein A–Sepharose. Beads were washed with 0.5% Triton X-100/PBS, and bound proteins were resolved by SDS-PAGE and immunoblotted with mouse monoclonal anti-Myc or anti-HA antibodies (1:1,000). In other experiments, extracts (100 μg of protein) of HeLa cells transfected with Myc-maspardin or Myc-maspardinS109A were incubated for 1–2 h at 4°C with 5 μg of mouse monoclonal anti-Myc antibodies precoupled to agarose (Santa Cruz Biotechnology) and immunoblotted with anti-ALDH16A1 antibodies (15 μg/ml). Lastly, cerebral cortices from C57/Bl6 mice were homogenized in 0.5% Triton X-100/PBS with a Dounce homogenizer (50 strokes), passaged through a 25-gauge needle, and processed as for HeLa cells.
Protein content determination
Protein concentrations were determined using the Pierce BCA Protein Assay Reagent (Thermo Fisher Scientific, Rockford, IL, USA) with bovine serum albumin as the standard.
RT-PCR
Total RNA was extracted using the TRI Reagent (Invitrogen) from adult C57/Bl6 mouse cortices. RNA was reverse-transcribed (RT) with the Superscript III First-Strand cDNA synthesis system (Invitrogen) using a poly-T20 primer. Gene-specific polymerase chain reaction (PCR) was carried out in an MJ Research PTC-200 DNA Engine Thermal Cycler (Bio-Rad) using primers specific for maspardin (forward: 5’-TTT CCG GCA GAT TTT GGC TC-3’, reverse: 5’-GCG CAC TCT GAT CAA ACA CA-3’) or ALDH16A1 (forward: 5’-GGC CCA GAC GGG CTG TAT GAG TAT-3’, reverse: 5’-TAC CTG CAG GGT GTG GCC TT-3’). Reactions were subjected to an initial denaturing step of 2 min at 94°C, followed by 25 cycles of 45 s at 94°C, 90 s at 60°C, and 60 s at 72°C, with a final extension of 72°C for 10 min. Maspardin and ALDH16A1 cDNAs were used as controls.
Results
Immunodetection of the maspardin protein
We generated anti-peptide antibodies directed against the N terminus (b5704) and C terminus (c3945) of human maspardin. Lysates of untransfected HeLa cells and cells overexpressing full-length maspardin were immunoblotted with these antibodies (Fig. 1a, b). Maspardin was detected at ∼33 kDa on immunoblots of cell lysates from HeLa cells as well as cells overexpressing recombinant maspardin, consistent with its predicted size (Fig. 1a, b). Subcellular localization studies in HeLa cells using confocal microcopy revealed that maspardin was distributed throughout the cell yet appeared clustered in the perinuclear region (Fig. 1c, d). Importantly, the maspardin-immunoreactive signals were eliminated by preincubating antibodies with their respective immunogenic peptides (Fig. 1e–h).
Characterization of polyclonal anti-maspardin antibodies. a, b Lysates from mock-transfected (Untrans; 30 μg protein per lane) or maspardin-transfected (3 μg protein) HeLa cells were immunoblotted with antibodies against N- (a) or C-terminal (b) regions of maspardin. Migration of molecular mass standards (in kilodalton) are shown to the left. c, d HeLa cells were immunostained using N-terminal (c) or C-terminal (d) antibodies, with detection by confocal microscopy. e–h Immunodetection for both N- and C-terminal antibodies, respectively, could be eliminated on immunoblots (e, f) and immunostaining (g, h) by preadsorption with the antigenic peptides
Distribution of endogenous maspardin
To examine the subcellular distribution of maspardin using cellular fractionation techniques, we used techniques focusing on solubility differences (Fig. 2a) and differential sedimentation (Fig. 2b). Each fraction was resolved by SDS-PAGE and analyzed for the presence of maspardin and various marker proteins: PLC-γ, a cytoplasmic protein; calnexin, an integral membrane protein; lamin A + C, a nuclear envelope protein; and OPA1, a mitochondrial protein. Using the Qproteome cell compartment kit, maspardin was detected in both cytoplasmic and membrane fractions with undetectable levels in the nucleus (Fig. 2a). Differential centrifugation and immunoblot analysis demonstrated that maspardin attributed to the membrane fraction was most prominent in the P3 fraction (Fig. 2b). The P3 pellet comprises endoplasmic reticulum, Golgi, and plasma membranes as well as small vesicles but not mitochondria. As expected, the mitochondrial protein OPA1 was abundant in the P2 pellet but not the P3 pellet. In contrast to the integral membrane protein calnexin, which is enriched in the P3 but not the soluble S3 fraction, maspardin is distributed in both S3 and P3 fractions (Fig. 2b).
Subcellular distributions of maspardin in HeLa cells. a Fractionation of HeLa cell lysates was performed using the Qiagen Qproteome Cell Compartment Kit. Fractions (20 μg protein per lane) were immunoblotted with antibodies against maspardin (no. c3945), the cytoplasmic protein PLC-γ, the endoplasmic reticulum protein calnexin, and the nuclear envelope protein lamin A + C. Abbreviations: C, cytosolic fraction; M, membrane fraction; N, nuclear fraction. b Subcellular fractions were prepared from HeLa cells by differential centrifugation. Fractions (30 μg of protein per lane) were immunoblotted with antibodies against maspardin, the mitochondrial protein OPA1, calnexin, and PLC-γ. Specific fractions are described in the “Materials and methods.” Abbreviation: H, homogenate. c Detergent-solubilized extracts prepared from HeLa cells were subjected to FPLC gel-exclusion chromatography, and aliquots of collected fractions were immunoblotted with anti-maspardin antibodies. Elution peaks for marker proteins (in kilodalton) are indicated across the top
Proteins that harbor α/β hydrolase fold domains have been found to function as monomers, homodimers, and homotrimers as well as heterodimers with other proteins in vivo [15–19]. To determine whether maspardin functions as a monomer or as part of a larger oligomeric complex, we used gel-exclusion FPLC. Detergent-solubilized maspardin eluted in a single peak at ∼33 kDa, most consistent with a native monomeric structure (Fig. 2c).
Subcellular localization of maspardin by immunocytochemistry
Given the localization of maspardin to membrane as well as cytosolic fractions, we investigated the subcellular localizations of maspardin in HeLa cells using confocal immunofluorescence microscopy. Consistent with results presented in Fig. 1, endogenous maspardin was distributed throughout the cytoplasm, with a distinct enrichment in the perinuclear region (Fig. 3). Colocalization with the Golgi matrix protein GM130, a cis-Golgi marker, and EEA1, a marker for early endosomes, was limited (Fig. 3a, b). However, the trans-Golgi marker γ-adaptin and the late endocytic compartment marker CD63 more significantly colocalized with maspardin (Fig. 3c, d). These results, consistent with an earlier report [10], suggest that maspardin may function at later stages of the endocytic sorting pathway rather than at earlier stages, Indeed, when we compared maspardin localization with LAMP1 and the mannose-6-phospate receptor, markers for late endosomes/lysosomes, partial colocalization was revealed for both (Fig. 3e, f). At the same time, we cannot rule out the possibility that some of the colocalization with these markers is fortuitous due to the predominant cytoplasmic distribution of maspardin.
Maspardin interaction with the aldehyde dehydrogenase ALDH16A1
Maspardin has been shown to bind CD4, and it has been proposed to function as a negative regulatory factor in CD4-dependent T cell activation [10]. However, maspardin is ubiquitously expressed, implying that it interacts with other proteins in CD4-negative cell types. We searched for other maspardin-interacting proteins in a non-CD4-expressing cell type, HeLa cells that express maspardin highly. Cell extracts from HeLa cells were coimmunoprecipitated using the C-terminal antibody c3945, and maspardin-associated proteins were identified by MALDI-TOF mass spectrometry. To minimize the possibility of artifactual binding of proteins to the resin, we precleared the cell extract with protein A–Sepharose beads and additionally used a rabbit polyclonal IgG immunoprecipitation as a negative control. Immunoprecipitates were resolved by SDS-PAGE, and only bands absent from IgG and anti-maspardin-negative control immunoprecipitates, or else substantially increased in concentration in the anti-maspardin precipitate as compared with the controls, were collected for MALDI-TOF mass spectroscopy identification. Two proteins clearly met such criteria (Fig. 4). The first, ∼33 kDa, was identified as maspardin (12.01% coverage, seven distinct peptides). The second was human ALDH16A1 (11.60% coverage, 12 distinct peptides), an aldehyde dehydrogenase family member of unknown function [20]. At least four other proteins were detected in maspardin immunoprecipitates as compared with IgG control (Fig. 4), but the validity of these has not yet been established via secondary confirmation.
Identification of maspardin-associated proteins in HeLa cells. HeLa cell lysates or else extraction buffer alone was immunoprecipitated using an anti-maspardin antibody (no. c3945), and in control experiments HeLa cells extracts were immunoprecipitated with nonimmune IgG. Proteins coimmunoprecipitating with maspardin were resolved on 10% SDS-PAGE gels and stained with MALDI-TOF-mass-spectroscopy-compatible Coomassie blue. Only proteins present specifically in HeLa + c3945, and not in the nonspecific binding control conditions, were excised for enzymatic digestion by trypsin and identification by mass spectrometry. Maspardin (∼33 kDa, arrow) and ALDH16A1 (∼80 kDa, arrow) were clearly identified. Other coimmunoprecipitating proteins are evident but have not been independently verified (arrowheads). Heavy and light chains of IgG are indicated
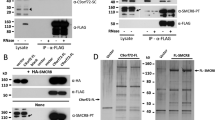
We considered the possibility that the coimmunoprecipitation of ALDH16A1 with maspardin was an artifact caused by cross-reactivity between the anti-maspardin antibodies and ALDH16A1 itself. However, this does not appear to be the case. When ALDH16A1 was overexpressed in HeLa cells, Western blot analysis with anti-maspardin antibodies revealed no immunoreactive signal at or near the predicted 85-kDa size of ALDH16A1 (data not shown). To validate further the ALDH16A1 interaction with maspardin, we overexpressed ALDH16A1 in HeLa cells. ALDH16A1 could be coimmunoprecipitated with endogenous maspardin regardless of which anti-maspardin antibody or which epitope-tagged ALDH16A1 was used (Fig. 5a, b). Furthermore, endogenous ALDH16A1 coimmunoprecipitated with overexpressed Myc-maspardin (Fig. 5c). Importantly, endogenous ALDH16A1 coimmunoprecipitated with endogenous maspardin (Fig. 5d). While endogenous ALDH16A1 could not be detected by immunoblotting in the input lane, following coimmunoprecipitation with anti-maspardin antibodies, ALDH16A1 was enriched and clearly became evident. This enrichment of ALDH16A1 following maspardin coimmunoprecipitation occurred following precipitations using both C- and N-terminal antibodies (Fig. 5d). Consistent with these interaction studies, examination of the subcellular localization of endogenous ALDH16A1 identified possible regions of punctuate colocalization with endogenous maspardin (Fig. 5e).
Maspardin associates with ALDH16A1 in HeLa cells. a, b HeLa cells were mock-transfected or else transfected with full-length ALDH16A1, epitope-tagged as indicated. Extracts were immunoprecipitated with control IgG (-IP) or polyclonal anti-maspardin antibodies to either the N-terminal (b5704) or the C-terminal (c3945) region and then immunoblotted with anti-Myc (a) or anti-HA (b) monoclonal antibodies to detect ALDH16A1. c Reciprocal immunoprecipitation experiments in which endogenous ALDH16A1 was immunoprecipitated using polyclonal anti-ALDH16A1 antibodies and then immunoblotted with anti-Myc antibodies to detect overexpressed Myc-maspardin. d Untransfected HeLa cell lysates or else control lysis buffer were immunoprecipitated with control IgG (-IP) or polyclonal anti-maspardin antibodies and immunoblotted with polyclonal anti-ALDH16A1 antibodies. e Confocal fluorescence microscopy of endogenous ALDH16A1 and maspardin demonstrate similar subcellular localizations within HeLa cells and distinct regions of colocalization (merge). Immunoreactive signals were greatly reduced by preadsorption of the antibodies with their respective antigenic peptides. Bars, 10 μm
Our extensive immunoprecipitation experiments certainly suggested a physiologically relevant maspardin–ALDH16A1 interaction. We next set out to determine whether this interaction was direct using an in vitro GST pull down binding assay. Full-length maspardin was expressed as a GST fusion protein and purified. In addition, full-length ALDH16A1 was expressed as a CBP fusion protein and purified. GST–maspardin or GST alone was immobilized on glutathione–Sepharose beads and incubated with purified CBP–ALDH16A1, then immunoblotted with anti-CBP antibodies. CBP–ALDH16A1 specifically bound to GST–maspardin but not to GST alone (Fig. 6a), indicating that the interaction is direct. Previously, it was demonstrated that a S109A mutation in maspardin prevented its interaction with CD4, presumably because Ser109 is part of the structural element resembling the nucleophile elbow of α/β hydrolases, and this fold domain serves a noncatalytic peptide-binding function [10]. To examine the role of this residue in the maspardin–ALDH16A1 interaction, wild-type and S109A mutant Myc-maspardin were expressed in HeLa cells. Anti-Myc antibodies precipitated endogenous ALDH16A1 in both cases (Fig. 6b). As in Fig. 5d, ALDH16A1 could not be detected in the input sample but only after enrichment of ALDH16A1 following maspardin immunoprecipitation. In addition, immunoblot analyses of cells overexpressing wild-type and mutant maspardin following immunoprecipitation using anti-ALDH16A1 antibodies demonstrated that both forms of maspardin could be coprecipitated (data not shown). These results suggest that Ser109 of maspardin is not critical for the maspardin–ALDH16A1 interaction.
Specificity of maspardin–ALDH16A1 interactions. a Maspardin interacts with ALDH16A1 in vitro. Immobilized GST–maspardin or control GST were used in pull down assays with purified CBP–ALDH16A1. Bound proteins were resolved by SDS-PAGE and immunoblotted with antibodies against CBP. b Lysates from wild-type Myc-maspardin (WT) or Myc-maspardin-S109A-transfected HeLa cells were immunoprecipitated using anti-Myc antibodies or control mouse IgG and then immunoblotted for ALDH16A1. Endogenous ALDH16A1 is not visible in the input lanes at this exposure but is enriched in anti-Myc immunoprecipitates. c Extracts from untransfected or Myc-ALDH16A1-transfected cells were immunoprecipitated with control IgG or anti-maspardin antibodies (Maspardin IP) and immunoblotted with anti-ALDH1A1 antibodies. In control experiments; protein A–Sepharose beads (Beads) alone were added without immunoglobulins. Migrations of the ALDH1A1 protein and IgG heavy chain are indicated. Migrations of molecular mass standards (in kilodalton) are shown to the right. In all panels, inputs represent 10% of the total lysate
Since ALDH161A is a member of a large superfamily comprising at least 19 different non-P450 aldehyde dehydrogenases, we sought to determine whether the interaction between maspardin and ALDH161A was specific for the ALDH16A1 isoform or whether this interaction was more generally present with other aldehyde dehydrogenases. One of the most widely studied aldehyde dehydrogenases is ALDH1A1 [20–23]. Coimmunoprecipitation experiments were conducted using cell lysates from mock-transfected or Myc-ALDH16A1-transfected HeLa cells, with immunoprecipitation using anti-maspardin antibodies followed by immunoblotting for ALDH1A1. In contrast to the ALDH16A1 isoform, ALDH1A1 is highly expressed endogenously in HeLa cells but was not detected following maspardin immunoprecipitation (Fig. 6c). These results suggest that the interaction between maspardin and ALDH16A1 is selective, though we cannot rule out interactions with other family members.
A compelling reason to investigate the function of maspardin is that loss-of-function mutations in this protein cause Mast syndrome, a complicated form of hereditary spastic paraplegia. Interestingly, Lein et al. [24] have reported that ALDH16A1 is expressed in the adult mouse brain, with high expression levels in the cerebral cortex. To determine whether the ALDH16A1–maspardin interaction could have relevance to the pathogenesis of Mast syndrome, we confirmed ALDH16A1 and maspardin expression in mouse cortex using RT-PCR (Fig. 7a). In addition, the endogenous interaction of maspardin and ALDH16A1 we first noted in HeLa cells also occurs in cerebral cortex since anti-maspardin antibodies coprecipitate ALDH16A1 (Fig. 7b). Lastly, when we investigated maspardin and ALDH16A1 localization patterns in mouse cerebral cortical neurons in situ, distinct puncta of colocalization were observed, most prominently in neuronal cell bodies and growth cones (Fig. 7c).
Maspardin and ALDH16A1 interactions in the central nervous system. a RT-PCR reaction products from total RNA extracted from mouse cortex. Sizes are 511 bp for maspardin and 512 bp for ALDH16A1. Full-length cDNA of maspardin and ALDH16A1 serve as both positive and negative controls, as shown. b Untransfected HeLa cell and mouse brain tissue lysates were immunoprecipitated with control IgG (for brain lysates only) or anti-maspardin antibodies and then immunoblotted with anti-ALDH16A1 antibodies. Where indicated (Buffer), no cell extracts were added. c Cultured primary cortical neurons were costained with antibodies against maspardin (green) and ALDH16A1 (red) and imaged using confocal microscopy. Boxed areas in the phase-contrast superimposed image on the left are enlarged on the right
Discussion
A mutation in the ACP33/maspardin gene results in a shift of reading frame and premature stop codon, causing maspardin protein truncation and presumed loss of function that results in the complicated form of hereditary spastic paraplegia known as Mast syndrome (SPG21). Maspardin was first identified as a CD4-binding protein and proposed as a negative regulatory factor involved in CD4-dependent T cell activation [10]. However, maspardin is a ubiquitously expressed protein, including CD4-negative cells types, and thus likely has other functions. In this study, we have characterized biochemical properties and localizations of endogenous maspardin and identified the aldehyde dehydrogenase ALDH16A1 as a binding partner.
Subcellular fractionation studies revealed a distribution of maspardin to both cytoplasmic and membrane compartments. Immunocytochemical localization studies demonstrated enrichment within the perinuclear region of the cell and colocalization with markers for trans-Golgi (γ-adaptin), late endosomes (CD63), and lysosomes (LAMP1), consistent with previous reports [10], supporting a functional role for maspardin in the trans-Golgi network/endosomal pathway. Several other proteins containing clusters of acidic amino acids, similar to the four Asp residues (residues 25–28) found near the N terminus of maspardin, are transported during intracellular trafficking events from various cellular compartments to the trans-Golgi network via interactions with sorting proteins known as phosphofurin acidic cluster sorting [25]. Interestingly, one of these transported proteins is the mannose 6-phosphate receptor, and our study demonstrates some colocalization of maspardin with the mannose 6-phosphate receptor. Therefore, maspardin may be involved in similar trafficking events. At the same time, some of the maspardin colocalization with these markers could reflect overlap with cytoplasmic maspardin staining.
We took an unbiased approach to identify novel maspardin-interacting proteins in HeLa cells, using immunoprecipitation analysis and subsequent MALDI-TOF peptide sequencing. We identified the aldehyde dehydrogenase ALDH16A1 as a maspardin-interacting protein and confirmed the interaction using coimmunoprecipitations of both endogenous and overexpressed proteins. Fusion protein pull downs further established that the interaction is direct. Immunofluorescence localization studies showed a widespread distribution of ALDH16A1 throughout the cytoplasm, with areas of colocalization with maspardin in both neurons and nonneuronal cells. Taken together, these studies support a physiologically relevant role for this interaction in vivo.
The human ALDH superfamily consists of at least 19 proteins with distinct chromosomal locations [20]. These enzymes catalyze the NAD(P)+-dependent irreversible oxidation of a wide spectrum of aliphatic and aromatic aldehydes during the metabolism of both endogenous and exogenous compounds [20]. Taken together, they are found in all subcellular regions including cytoplasm, mitochondria, endoplasmic reticulum, and nucleus, with some localized to more than one compartment. Unfortunately, nothing is known about ALDH16A1 function, and thus it remains unclear whether the interaction with ALDH16A1 influences the presumed functions of maspardin in the trans-Golgi network/endosomal pathway and what, if any, role ALDH16A1 plays in the pathogenesis of Mast syndrome. One possibility is that, in the absence of maspardin, ALDH16A1 is not trafficked to its proper location and cannot function, mimicking a loss of ALDH16A1 function. In fact, there is clear evidence that functional alterations of other ALDH proteins cause neurological disease. Mutations in ALDH3A2 result in Sjögren–Larsson syndrome, a neurological disorder characterized by mental retardation, congenital ichthyosis, and spastic paraparesis [26, 27]. Additionally, mutations in ALDH1A2, ALDH4A1, ALDH5A1, ALDH6A1, and ALDH7A1 lead to various neurological impairments and/or neurodevelopmental malfunctions [28–32], and a polymorphism of the ALDH2 gene may be a risk factor for late-onset Alzheimer’s disease [33].
How could aldehyde dehydrogenases in general be involved in neuronal dysfunction and death? One possibility is that long-lived aldehydes are highly reactive, and they can form adducts with various cellular targets, leading to impairments in cellular processes and even cell death [20]. Since aldehyde dehydrogenases are enzymes that participate in aldehyde detoxification by catalyzing their oxidation, a loss of ALDH16A1 localization or enzymatic function in patients with SPG21 may signify a particular vulnerability for the upper motor neurons. Interestingly, a recent study reported that the autosomal-recessive pure hereditary spastic paraplegia SPG5 (MIM 270800) results from loss-of-function mutations in the cytochrome P450 enzyme CYP7B1, a 7α-hydroxylase that is key for metabolism of brain cholesterol and neurosteroids [34]. Further studies of the function of ALDH16A1 and the maspardin–ALDH16A1 interaction in neuronal cells may help clarify the cellular pathogenesis underlying Mast syndrome.
References
Schwarz GA, Liu CN (1956) Hereditary (familial) spastic paraplegia: further clinical and pathologic observations. AMA Arch Neurol Psychiatry 75:144–162
Harding AE (1983) Classification of the hereditary ataxias and paraplegias. Lancet 1:1151–1155. doi:10.1016/S0140-6736(83)92879-9
Crosby AH, Proukakis C (2002) Is the transportation highway the right road for hereditary spastic paraplegia? Am J Hum Genet 71:1009–1016. doi:10.1086/344206
Reid E (2003) Science in motion: common molecular pathological themes emerge in the hereditary spastic paraplegias. J Med Genet 40:81–86. doi:10.1136/jmg.40.2.81
Fink JK (2006) Hereditary spastic paraplegia. Curr Neurol Neurosci Rep 6:65–76. doi:10.1007/s11910-996-0011-1
Soderblom C, Blackstone C (2006) Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol Ther 109:42–56. doi:10.1016/j.pharmthera.2005.06.001
Züchner S (2007) The genetics of hereditary spastic paraplegia and implications for drug therapy. Expert Opin Pharmacother 8:1433–1439. doi:10.1517/14656566.8.10.1433
Stevanin G, Ruberg M, Brice A (2008) Recent advances in the genetics of spastic paraplegias. Curr Neurol Neurosci Rep 8:198–210. doi:10.1007/s11910-008-0032-z
Simpson MA, Cross H, Proukakis C, Pryde A, Hershberger R, Chatonnet A, Patton MA, Crosby AH (2003) Maspardin is mutated in mast syndrome, a complicated form of hereditary spastic paraplegia associated with dementia. Am J Hum Genet 73:1147–1156. doi:10.1086/379522
Zeitlmann L, Sirim P, Kremmer E, Kolanus W (2001) Cloning of ACP33 as a novel intracellular ligand of CD4. J Biol Chem 276:9123–9132. doi:10.1074/jbc.M009270200
Zhu P-P, Patterson A, Lavoie B, Stadler J, Shoeb M, Patel R, Blackstone C (2003) Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J Biol Chem 278:49063–49071. doi:10.1074/jbc.M306702200
Zhu P-P, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C (2004) Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem 279:35967–35974. doi:10.1074/jbc.M404105200
Blackstone C, Roberts RG, Seeburg DP, Sheng M (2003) Interaction of the deafness–dystonia protein DDP/TIMM8a with the signal transduction adaptor molecule STAM1. Biochem Biophys Res Commun 305:345–352. doi:10.1016/S0006-291X(03)00767-8
Zheng YL, Li BS, Veeranna, Pant HC (2003) Phosphorylation of the head domain of neurofilament protein (NF-M): a factor regulating topographic phosphorylation of NF-M tail domain KSP sites in neurons. J Biol Chem 278:24026–24032. doi:10.1074/jbc.M303079200
Schrag JD, Li YG, Wu S, Cygler M (1991) Ser-His-Glu triad forms the catalytic site of the lipase from Geotrichum candidum. Nature 351:761–764. doi:10.1038/351761a0
Holmquist M (2000) Alpha beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci 1:209–235. doi:10.2174/1389203003381405
Elmi F, Lee HT, Huang JY, Hsieh YC, Wang YL, Chen YJ, Shaw SY, Chen CJ (2005) Stereoselective esterase from Pseudomonas putida IFO12996 reveals alpha/beta hydrolase folds for d-beta-acetylthioisobutyric acid synthesis. J Bacteriol 187:8470–8476. doi:10.1128/JB.187.24.8470-8476.2005
Hecht HJ, Sobek H, Haag T, Pfeifer O, van Pee KH (1994) The metal-ion-free oxidoreductase from Streptomyces aureofaciens has an alpha/beta hydrolase fold. Nat Struct Biol 1:532–537. doi:10.1038/nsb0894-532
Chen CC, Han Y, Niu W, Kulakova AN, Howard A, Quinn JP, Dunaway-Mariano D, Herzberg O (2006) Structure and kinetics of phosphonopyruvate hydrolase from Variovorax sp. Pal2: new insight into the divergence of catalysis within the PEP mutase/isocitrate lyase superfamily. Biochemistry 45:11491–11504. doi:10.1021/bi061208l
Marchitti SA, Brocker C, Stagos D, Vasiliou (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4:697–720. doi:10.1517/17425255.4.6.697
Jacobs FMJ, Smits SM, Noorlander CW, von Oerthel L, van der Linden AJ, Burbach JP, Smidt MP (2007) Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 134:2673–2684. doi:10.1242/dev.02865
Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V (2007) Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem 282:25668–25676. doi:10.1074/jbc.M702076200
Vasiliou V, Nebert DW (2005) Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics 2:138–143
Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. doi:10.1038/nature05453
Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G (1998) PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94:205–216. doi:10.1016/S0092-8674(00)81420-8
Rizzo WB (2007) Sjögren–Larsson syndrome: molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol Genet Metab 90:1–9. doi:10.1016/j.ymgme.2006.08.006
Rizzo WB, Carney G (2005) Sjögren–Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2). Hum Mutat 26:1–10. doi:10.1002/humu.20181
Valle D, Goodman SI, Applegarth DA, Shih VE, Phang JM (1976) Type II hyperprolinemia: 1-pyrroline-5-carboxylic acid dehydrogenase deficiency in cultured skin fibroblasts and circulating lymphocytes. J Clin Invest 58:598–603. doi:10.1172/JCI108506
Gibson KM, Hoffmann GF, Hodson AK, Bottiglieri T, Jakobs C (1998) 4-Hydroxybutyric acid and the clinical phenotype of succinic semialdehyde dehydrogenase deficiency, an inborn error of GABA metabolism. Neuropediatrics 29:14–22. doi:10.1055/s-2007-973527
Vasiliou V, Pappa A (2000) Polymorphisms of human aldehyde dehydrogenases: consequences for drug metabolism and disease. Pharmacology 61:192–198. doi:10.1159/000028400
Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MAAP, Omran H, Tacke U, Uhlenberg B, Weschke B, Clayton PT (2006) Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med 12:307–309. doi:10.1038/nm1366
Deak KL, Dickerson ME, Linney E, Enterline DS, George TM, Melvin EC, Graham FL, Siegel DG, Hammock P, Mehltretter L, Bassuk AG, Kessler JA, Gilbert JR, Speer MC, NTD Collaborative Group (2005) Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2. Birth Defects Res A Clin Mol Teratol 73:868–875. doi:10.1002/bdra.20183
Kamino K, Nagasaka K, Imagawa M, Yamamoto H, Yoneda H, Ueki A, Kitamura S, Namekata K, Miki T, Ohta S (2000) Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late-onset Alzheimer's disease in the Japanese population. Biochem Biophys Res Commun 273:192–196. doi:10.1006/bbrc.2000.2923
Tsaousidou MK, Ouahchi K, Warner TT, Yang Y, Simpson MA, Laing NG, Wilkinson PA, Madrid RE, Patel H, Hentati F, Patton MA, Hentati A, Lamont PJ, Siddique T, Crosby AH (2008) Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet 82:510–515. doi:10.1016/j.ajhg.2007.10.001
Acknowledgments
The authors wish to thank Dr. Howard Jaffe (NINDS Protein/Peptide Sequencing Facility) for mass spectrometry, James Nagle and Debbie Kauffman (NINDS DNA Sequencing Facility) for DNA sequencing, and Henri Jupille and Julia Stadler for technical assistance. This study was supported by the Intramural Research Program of the NIH, National Institutes of Neurological Disorders and Stroke.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hanna, M.C., Blackstone, C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics 10, 217–228 (2009). https://doi.org/10.1007/s10048-009-0172-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-009-0172-6